Part:BBa_K4391001
Penicillin G Aptamer
This is an aptamer that binds to Penicillin G or Benzyl Penicillin. The aptasensor allows the detection of penicillin in a wide concentration range, comprised between 0.4 and 1000 µg L−1. The aptamer folded structure of the aptamer was modelled and the aptamer was docked with it's target Penicllin G molecule. The aptamer was also electrochemically characterized after fixing onto GO/PANI.
Usage and Biology
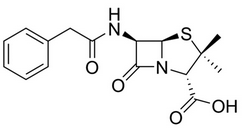
In veterinary practice, benzylpenicillin or penicillin G (G for gold standard) is the most frequently used β-lactam antibiotic for prevention and treatment of bacterial infection like mastitis for dairy cattle. Penicillin G present in milk might be responsible of allergic reaction in sensitized individuals. Penicillin residues may also be responsible for the induction of antibiotics resistant bacteria and in other level can negatively affect the growth of starter cultures for fermented dairy products. Thus a quick and cheap method of Penicillin G detection is imperative. The 2D and 3D structures of the Penicillin G or Benzyl Peniciilin moiety are shown in Fig-1 and Fig-2 respectively.
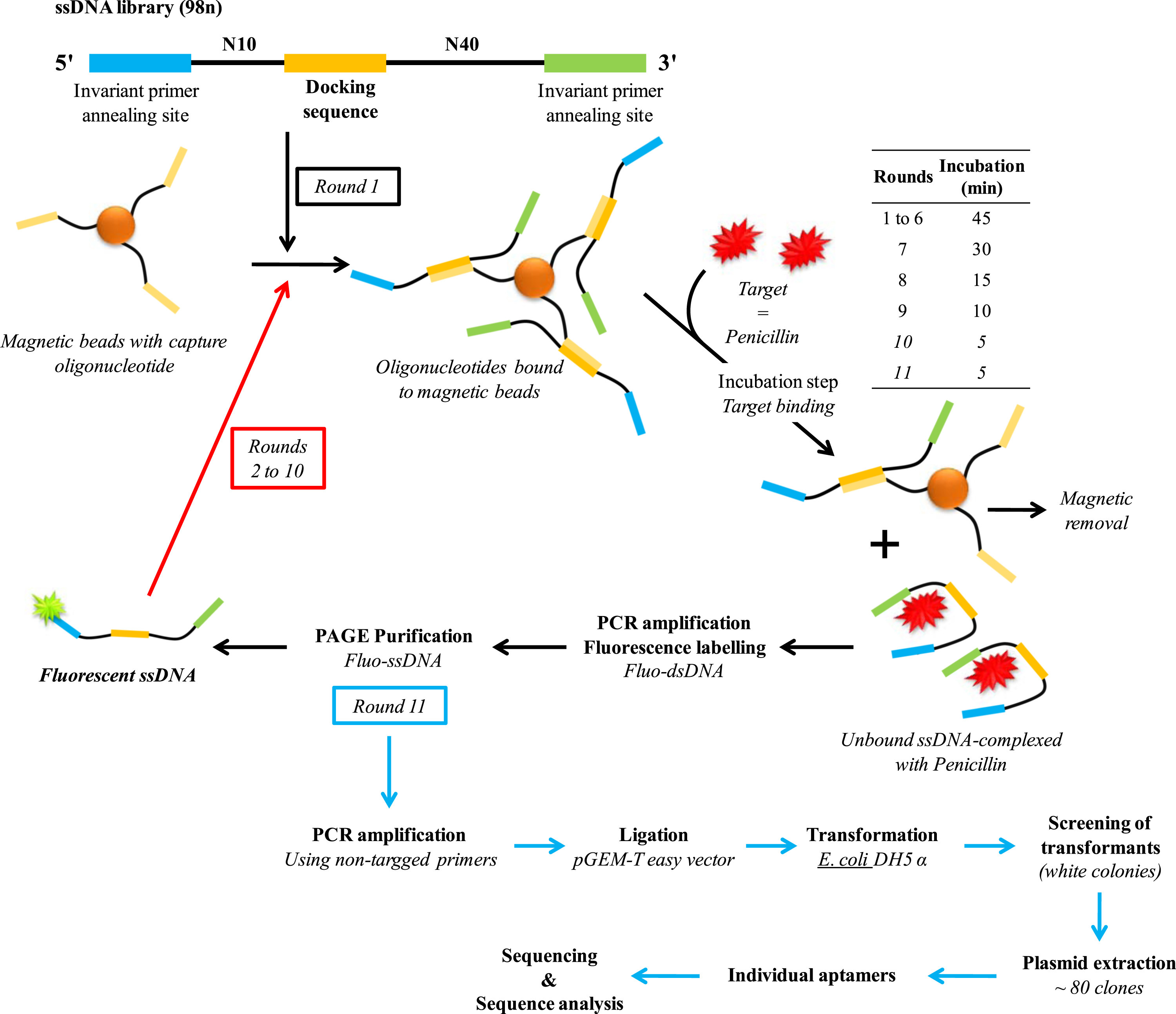
Aptamers are oligonucleotides (DNA or RNA) that can bind with high affinity and specificity to various targets, including a large number of molecules containing randomly created sequences, such as drugs, proteins or other organic or inorganic molecules. Owing to their many advantages such as cost effectiveness, flexibility, easy of modification, high stability, regeneration capacity, and compatibility with large-scale production, aptamers are a first choice candidate for the determination of penicillin in dairy products. This aptamer sequence was selected by Paniel et. al. using a particular SELEX (systematic evolution of ligands by exponential enrichment) process called Capture-SELEX (shown in Fig-3).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Modelling
(Section added by iGEM 2022 team IISER_Mohali)
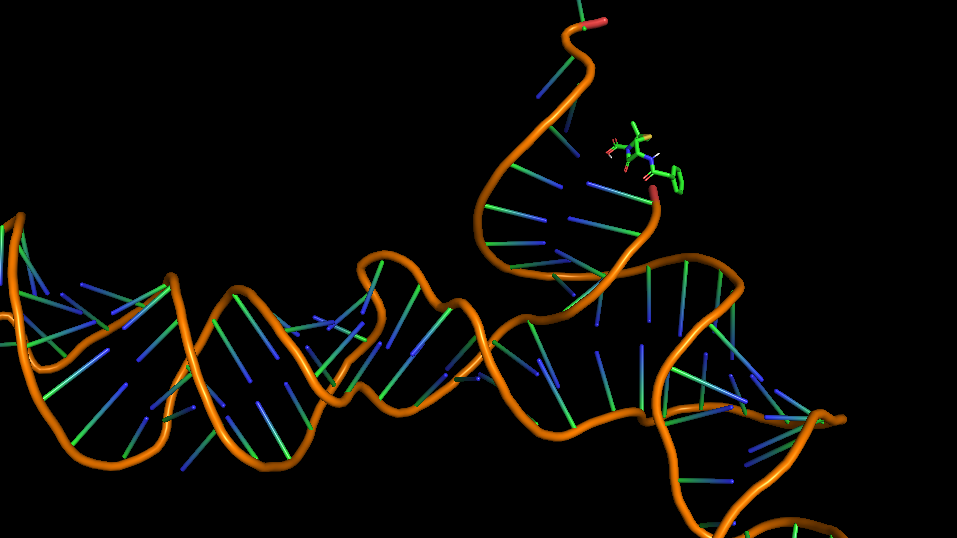
3D structure of the Aptamer
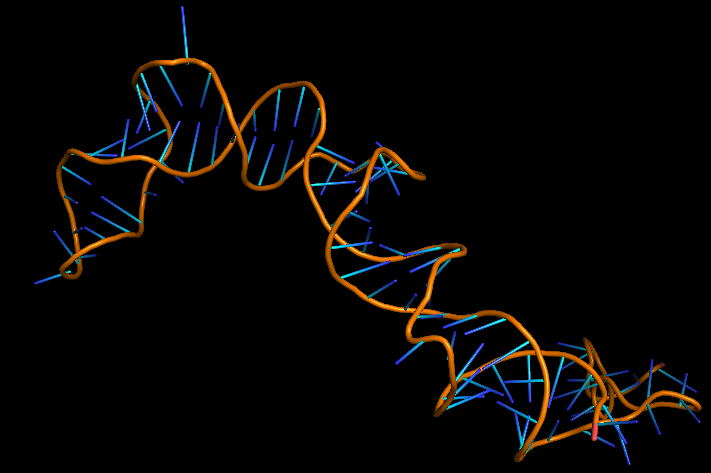
The 3D folded structure of the aptamer was generated using Xiao Lab 3dRNA/DNA - An RNA and DNA tertiary structure prediction method (http://biophy.hust.edu.cn/new/3dRNA/create). The input was the DNA aptamer sequence, molecule type DNA, Predict 2nd Structure: Yes, 2nd Structure Prediction Method: RNA Fold, Procedure Best, # of Predictions 5. No changes were made in Advanced options. Model-Fig-1 shows the most stable folded 3D structure.
Molecular Docking
Penicillin G or Benzylpenicillin was docked with it's aptamer using AutoDock Version 4.2. The main steps involed in the docking process:
- Adding polar hydrogen and kollman charges to the receptor and saving it as pdbqt
- Defining torsion root and torsion angles for ligand and saving it as pdbqt
- Making grid box (including whole of the protein) and defining grid parameters for receptor using autogrid
- Docking using Genetic algorithm (ga runs: 50, population size:300, evals: 250,000; generations: 27000)
- Output: Lamarckian GA
- Run using autodock
- Analysis of results using the dlg output file
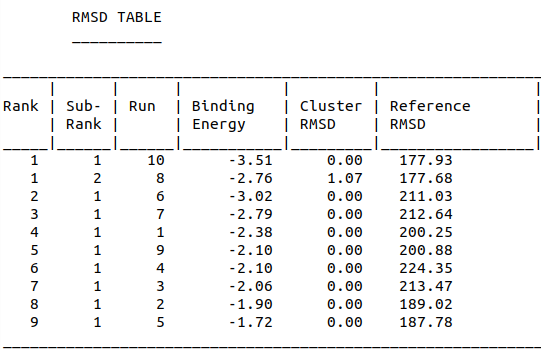
Model-Table-1 shows binding energy for the top 10 docked structures, while Model-Table-2 shows the RMSD scores for the top 10 docked structures. We see that the docked configuration corresponding to run 10, is the most probable one (least energy and highest RMSD score). This is shown in Model-Fig-2.
Characterization
We ordered our DNA aptamers from IDT as 2 μmole DNA oligo in standard desalting conditions. We resuspended the lyophilized aptamer with 0.1 molar PBS. We measured the Cyclic voltammetry and impedance of the bare Glassy Carbon electrode before modifying it with GO/PANI solution, and then after fixing Penicillin-G aptamer on it.
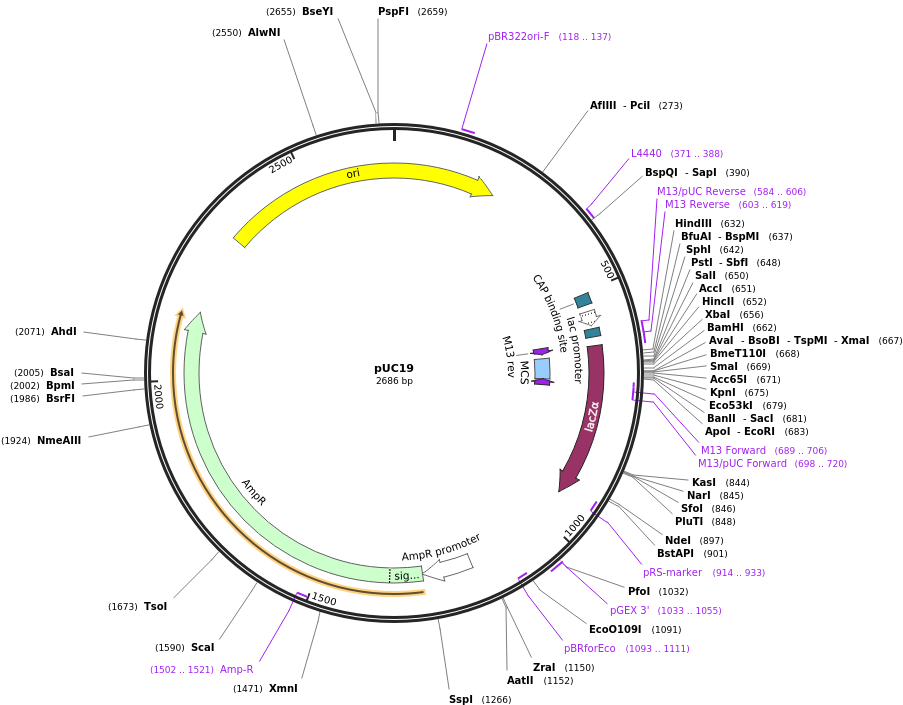
We have also cloned the aptamer into pUC19 vector (shown in Char-Fig-1) by restriction digestion using Xba1 and BamH1. This was followed by transformation into Escherichia coli DH5α.
Fixing of Aptamers on GO/PANI and electrochemical characterization
- Graphene oxide (GO) was prepared using a modified Hummer method.
- A conducting polymer Polyaniline (PANI) was mixed into a GO in order to improve its functionalities and charge conduction value that can form a suitable interaction with the aptamer molecule.
- GO/PANI was electrodeposited onto a glassy carbon (GC) electrode.
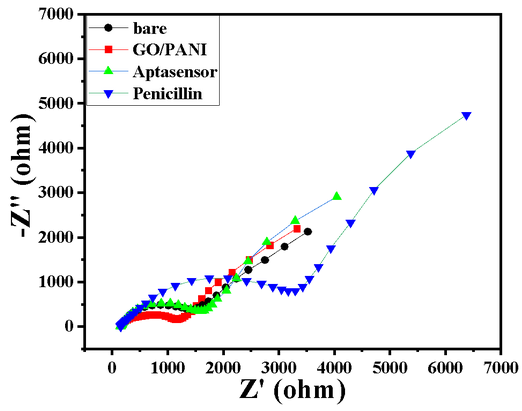
- A change in the RCT from 1400 Ω to 1100 Ω after deposition shows the GO/PANI layer formation on the GC surface, shown in Char-Fig-2
- The Cyclic Voltammetric (CV) and Electrochemical impedance spectroscopy (EIS) response of GC was measured in 1 mM Fe3+/Fe2+ solution in 0.1M PBS.
- EIS was measured in the frequency range from 100 kHz to 10 mHz, ∆E value of the CV is shown by Char-Fig-3
- The lyophilized aptamer was diluted with 0.1M PBS solution to 250nM aptamer and was used for Self-assembled monolayer (SAM) formation.
- 10µL of aptamer solution was drop cast on the modified GC and was kept for drying overnight (12h) at 4 °C.
- The SAM aptasensor was then dropcast with the Penicillin G and kept to dry and its electrochemical response was measured.
- The Nyquist plot of the electrode where the aptamer molecule shows an increase in the RCT (Char-Fig-3) showing the successful assembly of the aptamer layer on the modified GC.
- It can be concluded from the RCT value and the size of the semi-circle of the Nyquist plot (Char-Fig-2) that sensitivity of the developed aptasensor towards penicillin is good.
References
[1] Nathalie Paniel, Georges Istamboulié, Athar Triki, Clément Lozano, Lise Barthelmebs, Thierry Noguer, Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor, Talanta, Volume 162, 2017, Pages 232-240, ISSN 0039-9140, https://doi.org/10.1016/j.talanta.2016.09.058. (https://www.sciencedirect.com/science/article/pii/S0039914016307287)
[2] Purkait, T., Singh, G., Kumar, D., Singh, M. & Dey, R. S. High-performance flexible supercapacitors based on electrochemically tailored three-dimensional reduced graphene oxide networks. Sci. Reports 2018 81 8, 1–13 (2018).
[3] Chergui, S., Rhili, K., Poorahong, S., & Siaj, M. (2020). Graphene Oxide Membrane Immobilized Aptamer as a Highly Selective Hormone Removal. Membranes, 10(9). https://doi.org/10.3390/membranes10090229
[4] Subramanian, P., Lesniewski, A., Kaminska, I., Vlandas, A., Vasilescu, A., Niedziolka-Jonsson, J., . . . Szunerits, S. (2013). Lysozyme detection on aptamer functionalized graphene-coated SPR interfaces. Biosensors and Bioelectronics, 50, 239-243. https://doi.org/https://doi.org/10.1016/j.bios.2013.06.026
[5] Zhao, Y., et al., Automated and fast building of three-dimensional RNA structures. Scientific Reports, 2012. 2: p. 734.
[6] Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S. and Olson, A. J. (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Computational Chemistry 2009, 16: 2785-91.
| None |