Part:BBa_K4192121
Plac-obcA-obcB-Plac-Fpoar, Oxalate secretion gene circuit
The expression products of obcA and obcB are enzymes needed to produce oxalate from citric acid and acetyl-CoA. And FpOAR protein is able to efflux oxalate. The composite part of these three genes can efflux oxalic acid out of the bacteria. We have already tested it in Pseudomonas fluorescens.
Characterization
CAU_China 2022 used this part as their oxalate secretion circuit. We transfered this circuit into pUC18 and pBBR1MCS2 to adapt to different chassis bacteria. This phenotype, in the way we test it, is reflected in the presence of more reducing substance outside the bacteria or changes in pH.
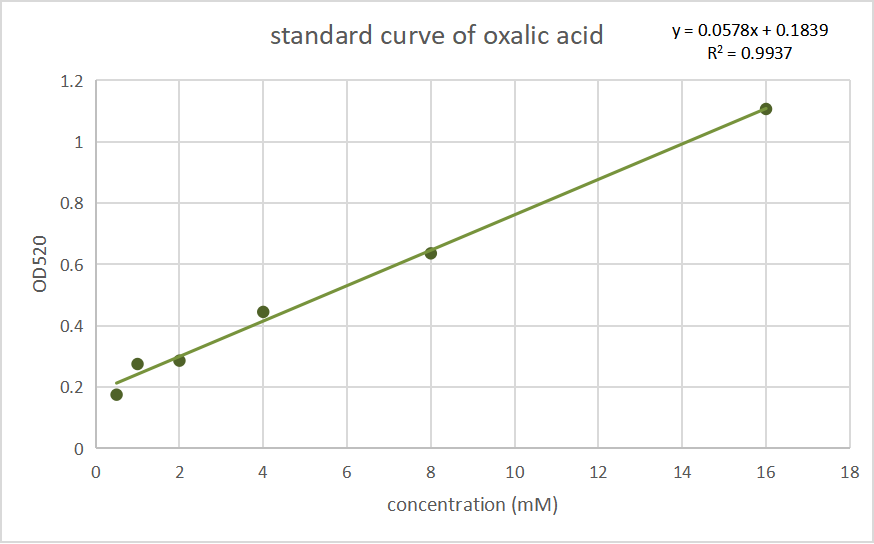
In P. fluorescens 2P24, we used the oxalic acid content detection kit (BoxBio, Beijing), which measure oxalic acid by salicylic acid complexed iron colorimetric method, allowing us to conduct colorimetric test for oxalic acid and other reducing substances in parallel treated samples with the same OD.
We added the same amount of induced bacterial solution to each of the medium, and they were first adjusted to the same OD595 with ddH2O. The control group is P. fluorescens 2P24 with an empty pBBR1MCS2 vector. After shaking culture at 28 ℃ for 12 hours, they were induced by IPTG. After another 8 hours, the bacterial solution was collected before we used a vacuum concentrator concentrate the products to ten times theirs original concentration.
Fig.1 Standard curves of oxalate concentration and absorbance at 520 nm.
Fig.2 Oxalic acid concentration test using 96-well plate. Lighter the red, higher the oxalic acid concentration in the well.
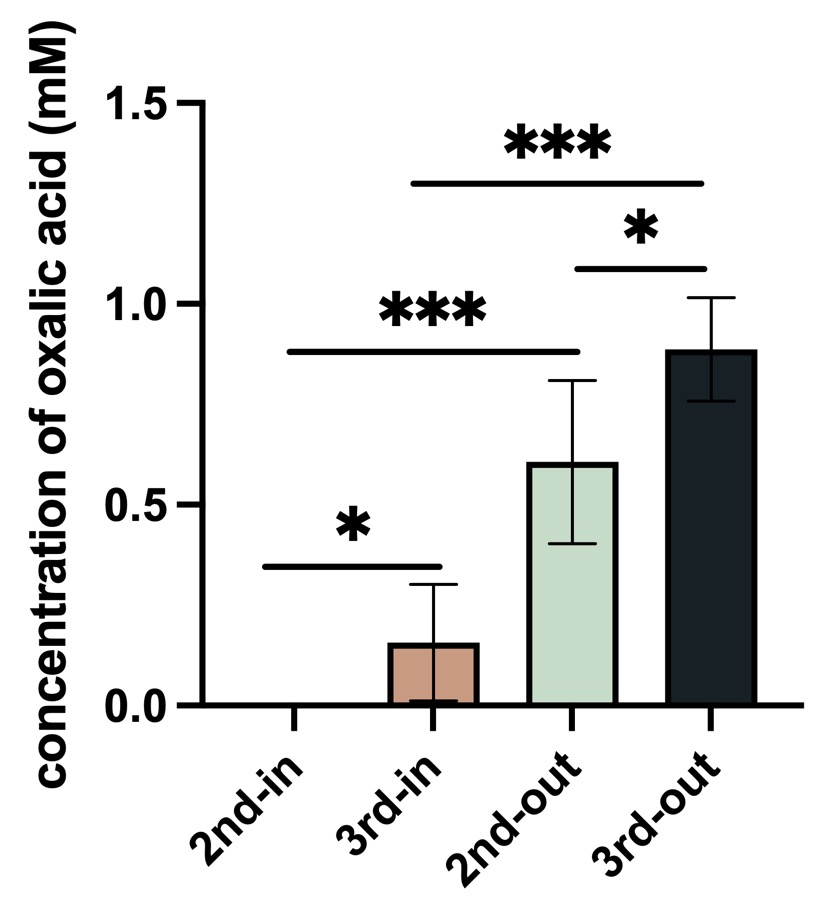
Fig.3 Result of oxalic acid concentration in P. fluorescens
Control: empty vector, 2nd: BBa_K4192120, 3rd:BBa_K4192121. In: Bacteria lysate, detected the oxalic acid concentration inside of the bacteria, out: the supernate of bacteria solution ,detected the oxalic acid concentration outside of the bacteria.
According to the results of ANOVA (Duncan's method), between these groups have a significant impact on the experimental results. We can come to the conclusion that the added Plac improved the efflux efficiency of oxalic acid, and this is our resulting oxalic acid producing bacteria.
In our project, this genetic circuit is used to form calcium oxalate protective film on the stone surface. We transferred it to Pseudomonas fluorescens and tested it on limestone surfaces.
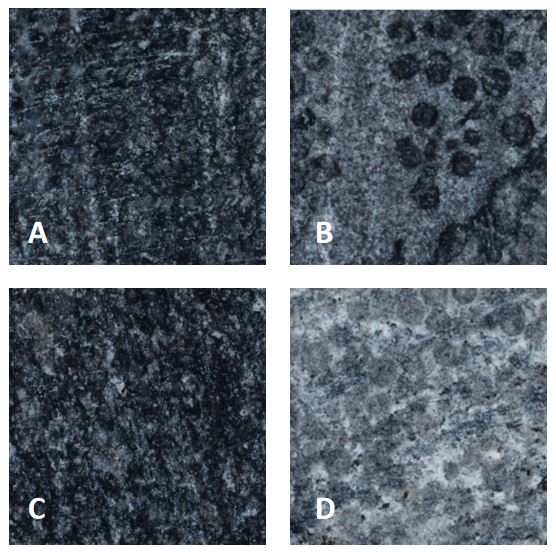
Fig.4 Scan results at 200 times magnification. A: LB control, B: chemical mineralization, C: untreated stone surface, D: BIMP.
A and C look exactly the same, showed black limestone surfaces with a small amount of white impurities. Both B and D produced white film material, but for the small circular depressions on the stone surface, the calcium oxalate formed by chemical mineralization did not cover it, while the biologic calcium oxalate film could obviously attach to it.
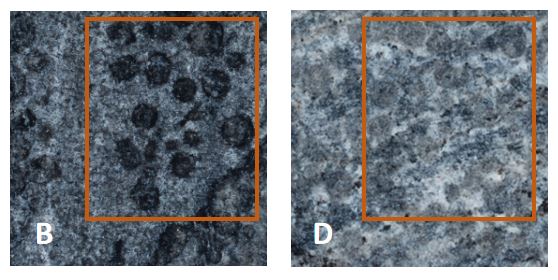
Fig.5 small circular depressions on the surface of B and D
This indicates that the calcium oxalate film formed by biological mineralization is beneficial to the protection of details. This is of great benefit to the preservation of artifacts with many carving details.
This is because the crystalline structures formed by BIMP are denser. And bacteria can enter tiny gaps, allowed BIMP occurring in tiny gaps. This conclusion can be drawn by observing and comparing crystals formed by different treatments: crystals formed by BIMP are clustered, with long morphology, that are not only closely arranged but also uniformity. Chemical mineralization, on the other hand, resulted in square, flat crystals that were loosely arranged and varied in thickness.
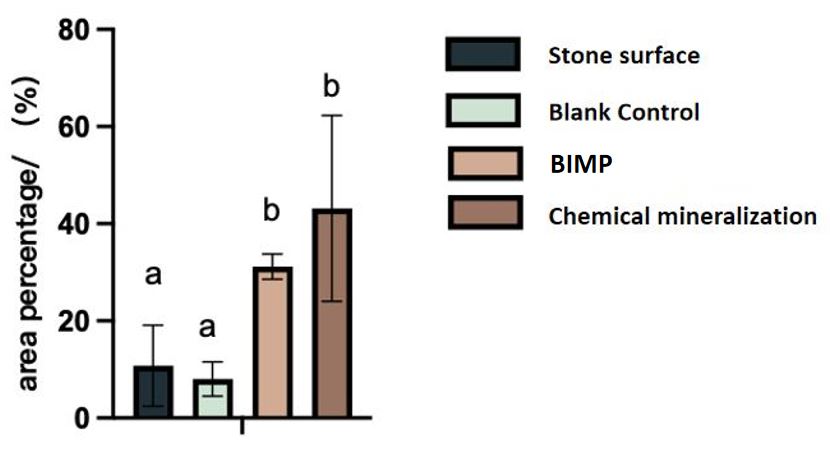
For the shake of interpret our conclusion more accurately, we randomly chose 5 areas on the scanned photos of stone surfaces, and used imageJ to conduct quantitative statistics on the white surface area.
Fig.6 The percentage of the white part on the stone surface. multiple hypothesis tests (Duncan's method) are conducted.
The same superscript letter means no difference, and different letters mean significant difference. That is, there is no difference between the LB treated blank control and the stone surface itself. Both BIMP and chemical mineralization lead to a significant increase in the amount of crystallization, while the yield of BIMP is more stable.
Here, we can draw a conclusion at the wet lab level: the engineered bacteria with oxalic acid secretion circuit can indeed conduct biolinalization on the stone surface, and the product has a dense structure to protect details. The BIMP products of our engineered bacteria are obvious, yield is stable, and compact enough to protect the details. This is enough to prove that P. fluorescens 2P24 with pBBR1MCS2-BBa_K4192121 has the rudiments of cultural relic protection ability. In this experiment, we only rely on the natural ability of Pseudomonas fluorescens to produce biofilm. We believe that if the production of biofilm is also enhanced, the protective effect of calcium oxalate film will be better.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2735
Illegal PstI site found at 397
Illegal PstI site found at 2878 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2735
Illegal PstI site found at 397
Illegal PstI site found at 2878
Illegal NotI site found at 2330 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2735
Illegal BglII site found at 1541
Illegal BglII site found at 3738
Illegal XhoI site found at 304
Illegal XhoI site found at 1428
Illegal XhoI site found at 3550 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2735
Illegal PstI site found at 397
Illegal PstI site found at 2878 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2735
Illegal PstI site found at 397
Illegal PstI site found at 2878
Illegal NgoMIV site found at 389
Illegal NgoMIV site found at 720
Illegal NgoMIV site found at 748
Illegal NgoMIV site found at 784
Illegal NgoMIV site found at 1014
Illegal NgoMIV site found at 1791
Illegal NgoMIV site found at 2208
Illegal AgeI site found at 326 - 1000COMPATIBLE WITH RFC[1000]
| None |