Part:BBa_K3823006
CstR
A transcription factor sensing hydrogen sulfide
Introduction
CstR is a CsoR-like sulfur transferase repressor found in Staphylococcus aureus[1], reported to react with HSnH and form sulfhydrated proteins, which can turn on their regulated genes[2] . Compared with other HSnH sensors, it shows significant response to HSnH induction. And it shows no response to H2O2, a structural analogue of HSSH and a common intracellular metabolite(Figure 1.).[3]

Results
1. To figure out how CstR works by sensing H2S, we characterized the function of CstR with Pcstr using pTrchis2A-CstR-Pcstr-mKate-CpSQR(donated by Professor Liu from Shandong University). The expression level regulated by Pcstr at different concentrations of S2- was shown by the fluorescence intensity of mKate(/OD600 nm).
At the beginning, bacteria cultured in 5 mL were treated with 0, 6.5, 13.0, 26.0 and 39.0 mg/L Na2S (0, 20, 40, 80 and 120 mg/L Na2S·9H2O) respectively, and then the fluorescence intensity of mKate was measured with a microplate reader(Figure 2.).
It can be seen that the presence of S2- at different concentrations has no significant effect on the growth of bacteria. However, with the increase of S2-, the fluorescence intensity increases at first and then decreases, indicating that when S2- is too high, the lifting effect on CstR inhibition is weakened. We hope to find out an appropriate concentration range in which the strength of Pcstr is positively correlated with the concentration of S2-, which tells us that we need to reduce the concentration gradient and concentration range for further characterization.
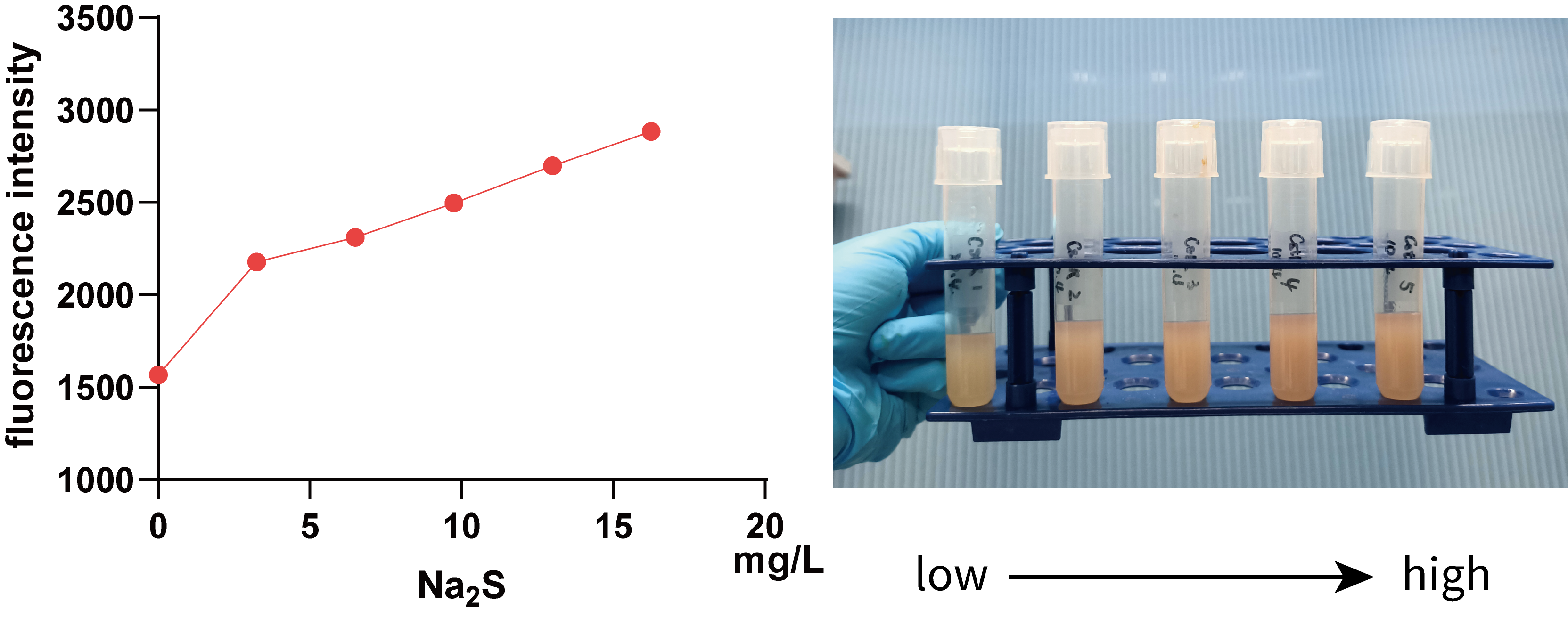
We further used 0, 3.25, 6.50, 9.75, 13.00, 16.25 mg/L Na2S (0, 10, 20, 30, 40 and 50 mg/L Na2S·9H2O) to treat 5 mL bacterial solution and then measured the fluorescence intensity of mKate using a microplate reader(Figure 3.).
It can be seen from the figure that in a smaller concentration range, there is a positive correlation between fluorescence intensity and Na2S concentration, visible to naked eyes, which implies that we can use data within this range to regulate Pcstr by giving different concentration H2S.
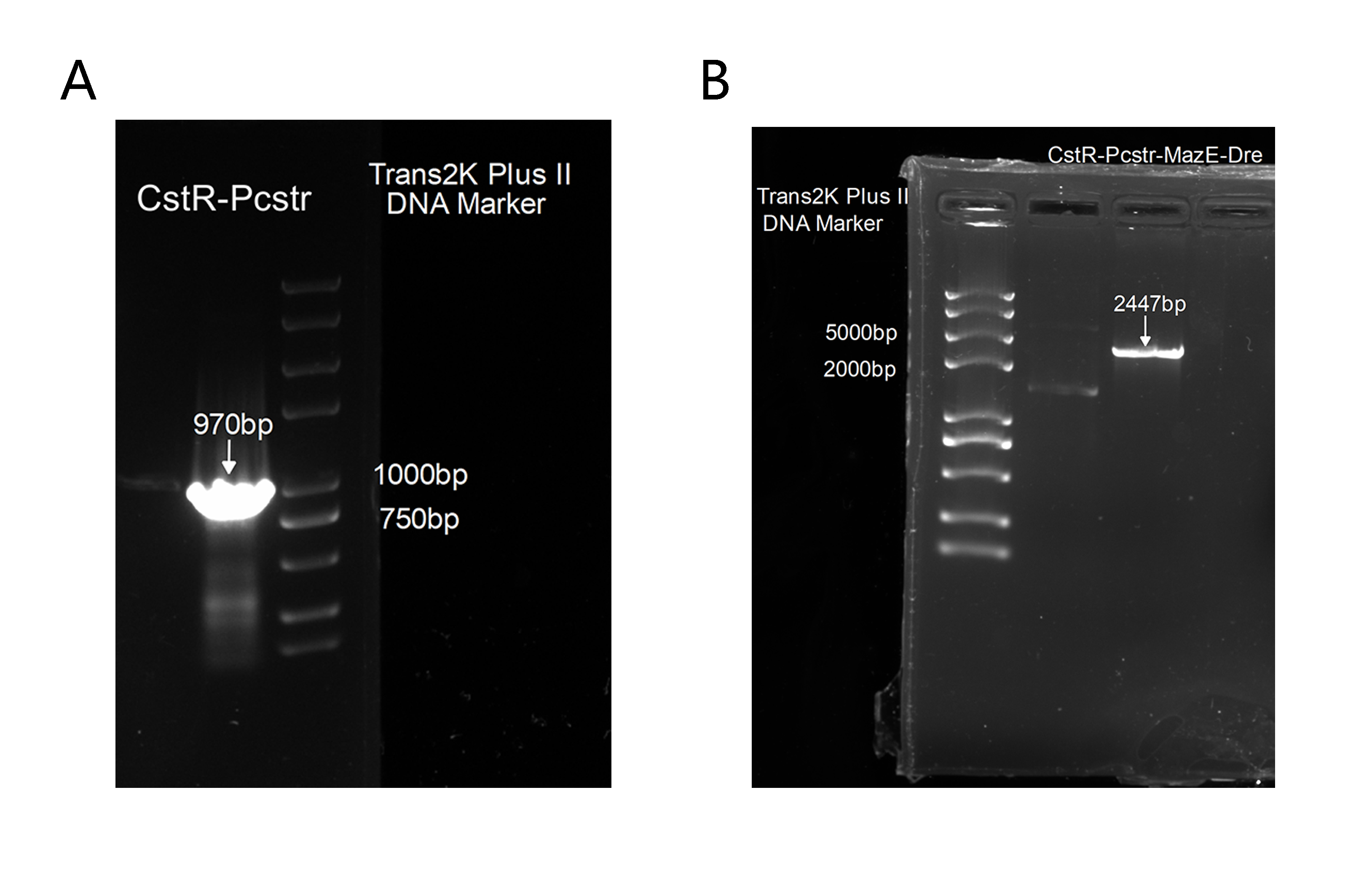
2. Also, to get this part for our plasmid construction, we successfully amplified CstR-Pcstr(Figure 3A), which was proved correct by sequencing and we has cloned it into our backbone to construct CstR-Pcstr-MazE-Dre(Figure 3B),which was also proved correct by sequencing.
References
- [1]Grossoehme, N., et al., Control of Copper Resistance and Inorganic Sulfur Metabolism by Paralogous Regulators in Staphylococcus aureus. Journal of Biological Chemistry, 2011. 286(15): p. 13522-13531.
- [2]Giedroc, D.P., A new player in bacterial sulfide-inducible transcriptional regulation. Mol Microbiol, 2017. 105(3): p. 347-352.
- [3]Liu, H., et al., Synthetic Gene Circuits Enable Escherichia coli To Use Endogenous H(2)S as a Signaling Molecule for Quorum Sensing. ACS Synth Biol, 2019. 8(9): p. 2113-2120.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 133
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 133
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 110
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 133
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 133
- 1000COMPATIBLE WITH RFC[1000]
| None |