Part:BBa_K3798032
Fusion protein sequence comprised with two CBD domain and one eGFP domain
The CBD domains of the protein is derived from the previous part of BBa_K1934020. The eGFP domain of the protein can be the signal of cellulose attachment and can be a subject of modeling. We use this protein to demonstrate the ability of using CBD domains to bind to the cellulose. By expressing this protein in E.coli in various concentration of iptg, we can analyze the fluorescent intensity to time, to model out the formation rate of the inclusion bodies.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Charcterization
Introduction
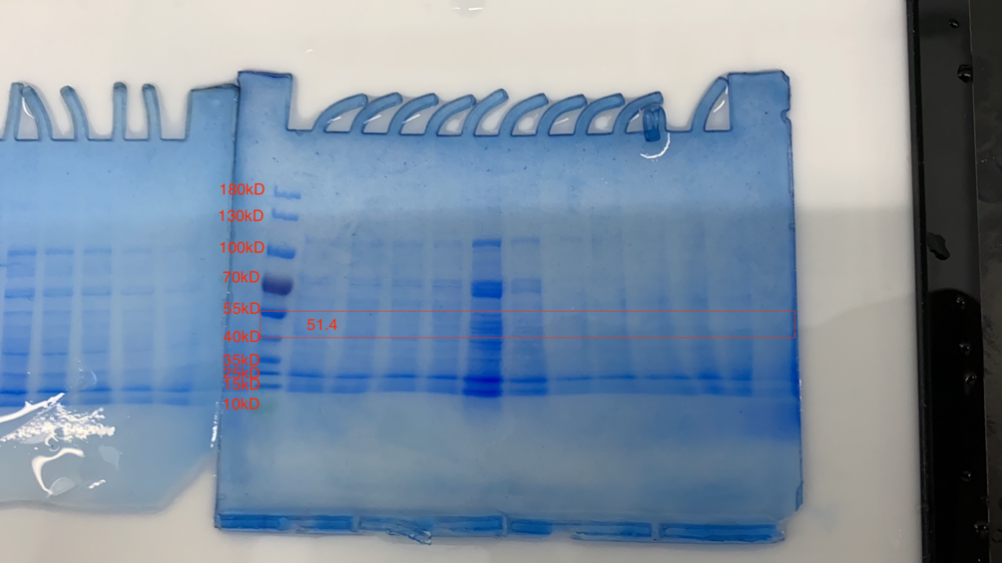
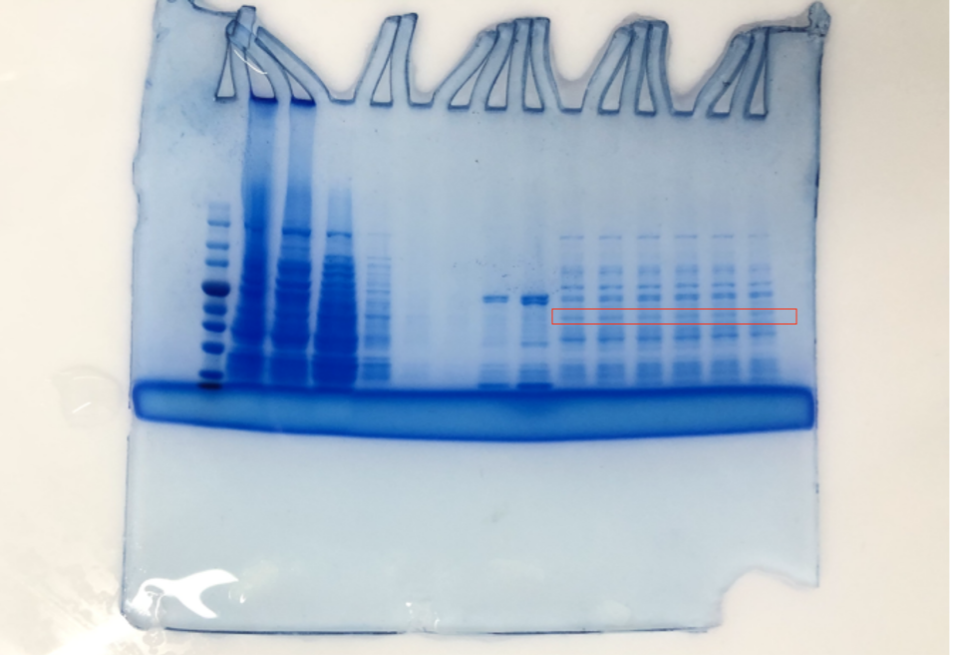
1.Cultivation of CBD-eGFP 1.1 Introduction In the experiments about the cultivation of fusion protein (CBD-SA), SHS1-pET28b-CBD-EGFP-CBD was inoculated into E.coil BL21. Then, the SHS1 proteins produced by E.coil BL21 were induced with IPTG in the culture medium. The target proteins were extracted using ultrasonic cell crusher. After purified the protein by using nickel column, we tested if these proteins have successfully expressed by the method of SDS-PAGE.
1.2 Failed attempt At the beginning of the experiments, we cultivated the proteins with carbon source of glucose and 0.3mM of IPTG to induce those proteins at 16℃.
Although the lysed cell is green indicating some protein had been expressed, the SDS-PAGE results of the purification does not yield satisfying results. As shown in the SDS gel above, no clear band of 51.4kDa is visible.
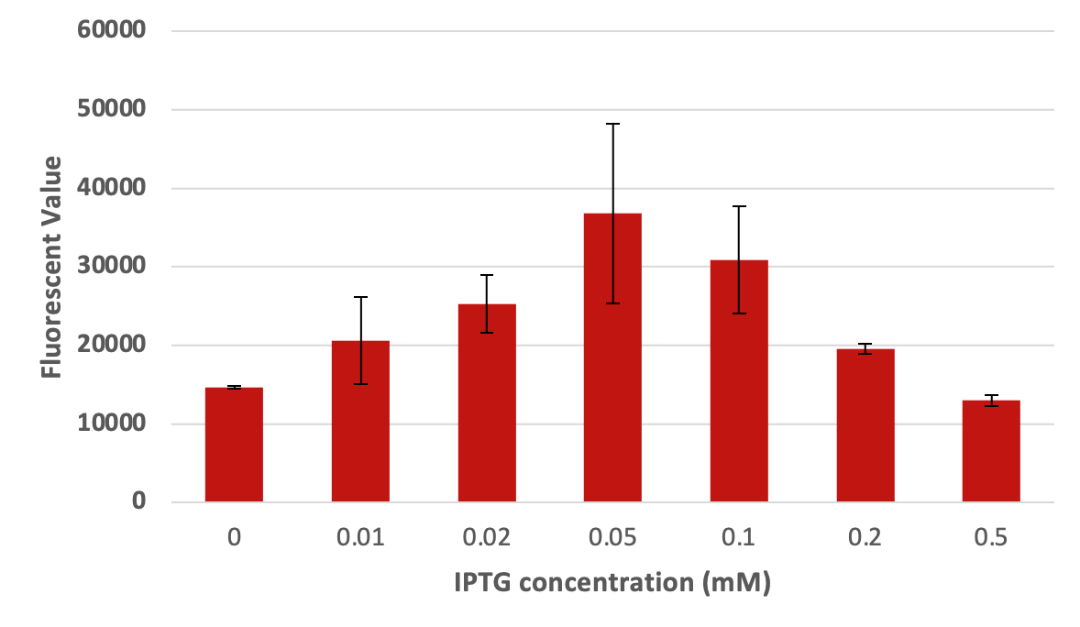
1.3 Learn and redesign To verify that the cause of experimental failure was inclusion body instead of incomplete lysed cells, we sonicated the remaining sediments of previous experiment with 300watts, which is as high as this machine can achieve that should be able to fully lysis any cells. However, after sonication, the supernatant of the sonicated cells still has relatively low fluorescence. The result might indicate that inclusion body formation is responsible for the failure of purification. Therefore, the following experiments are meanly focused on improving protein production by reducing the inducing intensity of the bacteria. We then tried to find out whether IPTG concentration can have effects on protein expression. We used different concentrations of IPTG to induce the expression of CBD-eGFP protein. From Figure3, it is obvious that the concentration of IPTG can have profound influences on the expression of the protein. Although it seems that IPTG concentration of 0.05mM and 0.1mM have the best expression result according to the following picture, we continued to proceed experiments to confirm the optimal IPTG concentration.
Concluded from the results of Figure 2, we hypothesized that the reason why our first attempt of expression failed was because of the IPTG concentration was too high.
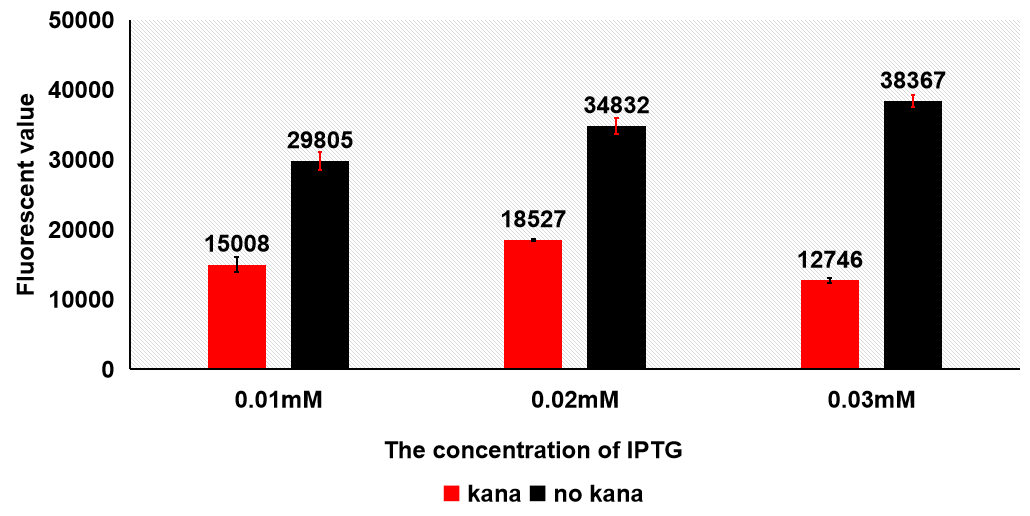
1.4 Antibiotics We used different concentration of IPTG to induce the protein at 16℃. The samples were divided into three groups based on the concentration of IPTG(0.01mM/0.02mM/0.03mM), and each group contains six samples which three of them were added kana while others were not. We tested the fluorescent value of all eighteen samples. The results were the samples which have no kana have higher fluorescent value, which means more protein are remained.
From this result, we decided to exclude kanamycin in the further experiment conditions.
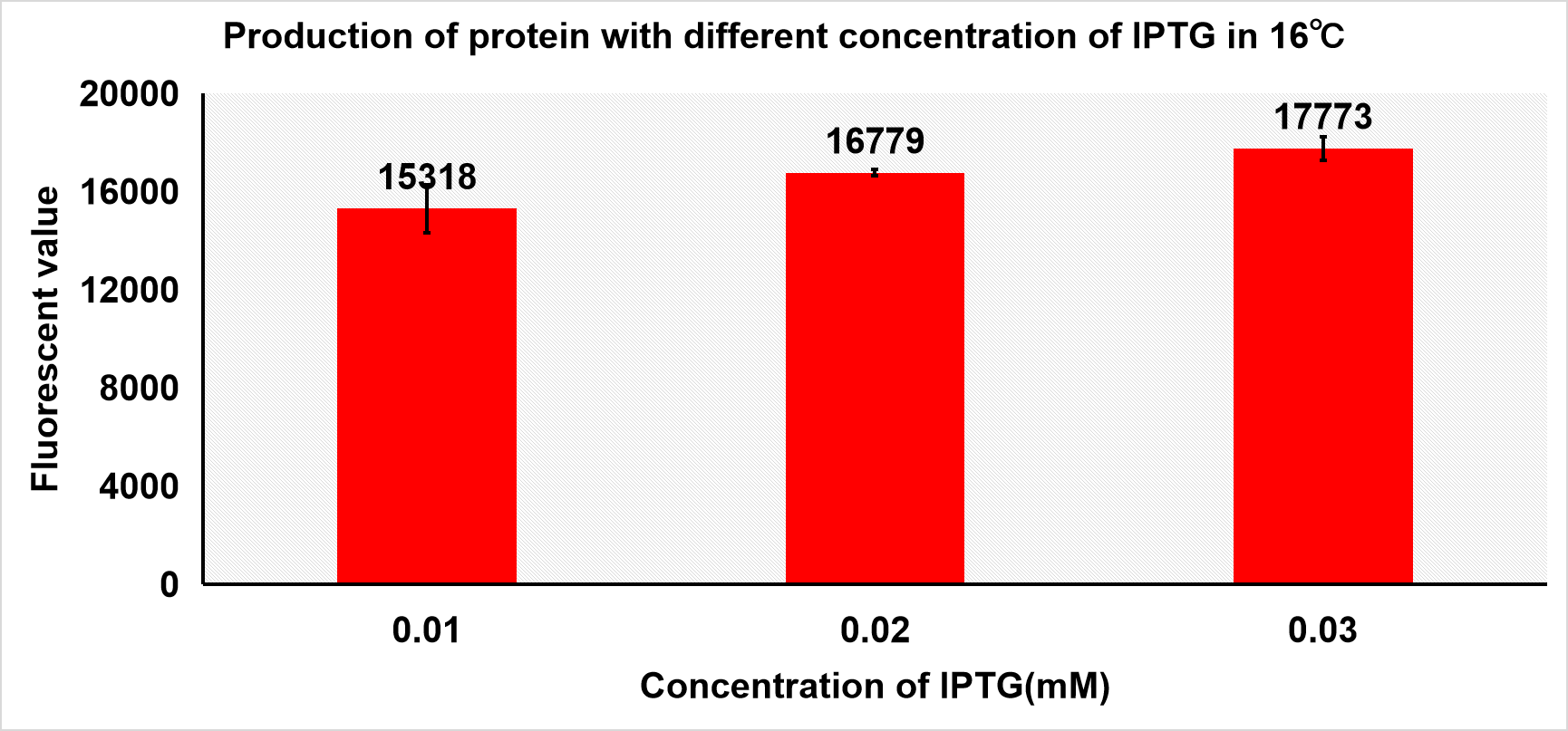
1.5 Optimization of experiment condition Then, we induced protein with different concentration of IPTG under 16 and 24℃ and both samples have no kana. Other conditions were the same.
As we can see in figure 3 a, the fluorescent value of the supernatant of sonicated cells at 16℃ of induction did not exceed 20000 in its best attempt of 0.03mM IPTG. If the fluorescent value of IPTG concentration is to increase linearly, then we can conclude that the production of protein is better under 24℃ since the 24℃ induced cells can yield a higher fluorescent value than that of the 16℃’s. Therefore, all the production attempts after this experiment utilize inducing temperature of 24℃. According to the fluorescent value, when the concentration of IPTG is 0.05mM and 0.2mM under 24℃, more protein was produced.
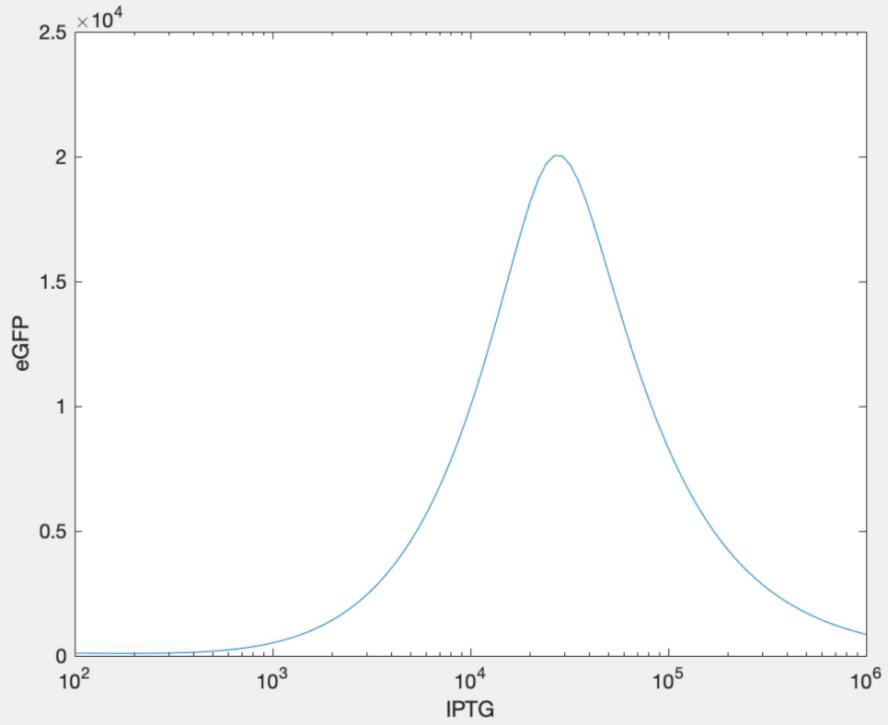
1.6 Modeling After all other factors except IPTG concentration are determined, we now aim to estimate the best IPTG concentration by mathematical simulation because it can provide a more accurate and rigorous concentration. We figured it out, through modeling, that when the IPTG concentration is 0.05mM, the protein expression can be maximized. For more information please click here: https://2021.igem.org./Team:SHSBNU_China/Model
After we got the results from modeling, we constructed a concentration gradient around 0.05mM and tried to find out the best condition for the induction of target proteins.
From the result above, it is obvious that the bacteria can produce the greatest amount of fusion protein with 0.05mM of IPTG. To our surprise, the best concentration was exactly 0.05mM, very consistent with our model. According to the result, the best condition to cultivated protein is 0.05mM IPTG at 24℃ without kanamycin. Thus, this value was going to be the optimal condition when we induced our protein.
We can obviously observe the expression of protein in the agarose gel, which means our experiment was successful.
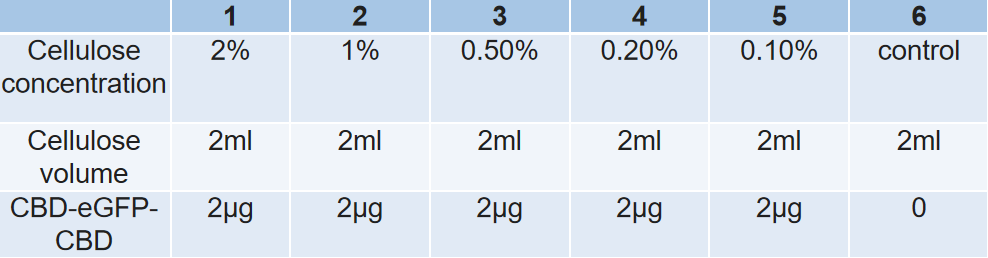
2.Connection between CBD-eGFP and cellulose 2.1 In this experiments, we hoped to find out the best condition for the binding between cellulose and CBD-SA. Firstly, we intended to find out how the concentration of cellulose affects the binding efficiency between cellulose and CBD-SA. According to the given cultivation environment above, 6 groups of different cellulose concentrations were set up, and the fluorescent value of each group was tested in the aim of calculating binding efficiency between Cellulose and CBD-SA.
Samples with different concentration of cellulose.
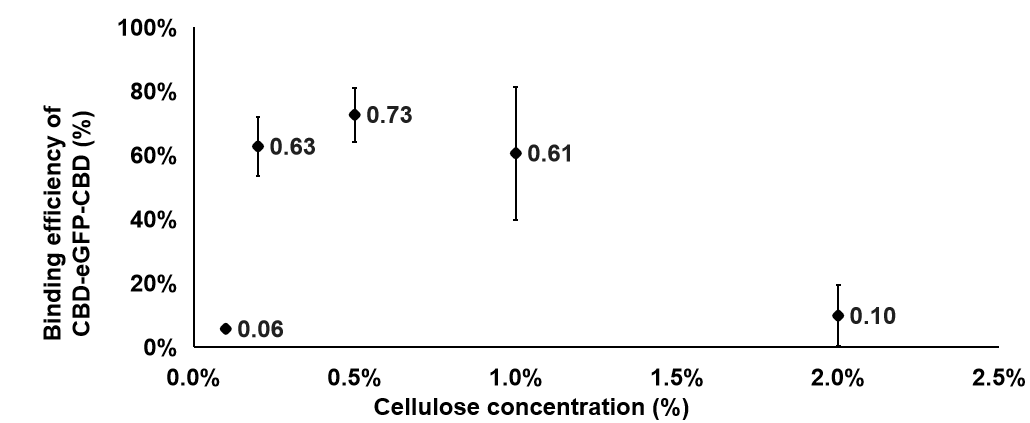
CBD-eGFP was used to replace CBD-SA in this experiment, because it has similar properties with CBD-SA and was designed as a visible fluorescent marker. Precipitate will form if cellulose binds the protein. That provided us a better way to exam the concentration of protein by detecting the fluorescent value. The fluorescent value of the supernatant in the solution was detected. The way we calculate the binding efficiency was as following: Firstly, we used the fluorescent value of supernatant in solution minus the fluorescence that already in the solution to get the remaining fluorescent value after the binding. Then, we used the total value minus the value we got and it refers to the binding value. The binding efficiency is the value which binding value divided by total fluorescence.
The formula is Binding efficiency = [Total fluorescence - (Remaining fluorescence - Background value)]/Total value
We can learn from the graph that the binding efficiency is relatively high when the concentration of cellulose is at 0.2% and 0.5%. Considering the cellulose will be sticky when the concentration is too high, we took 0.2% as our finally concentration.
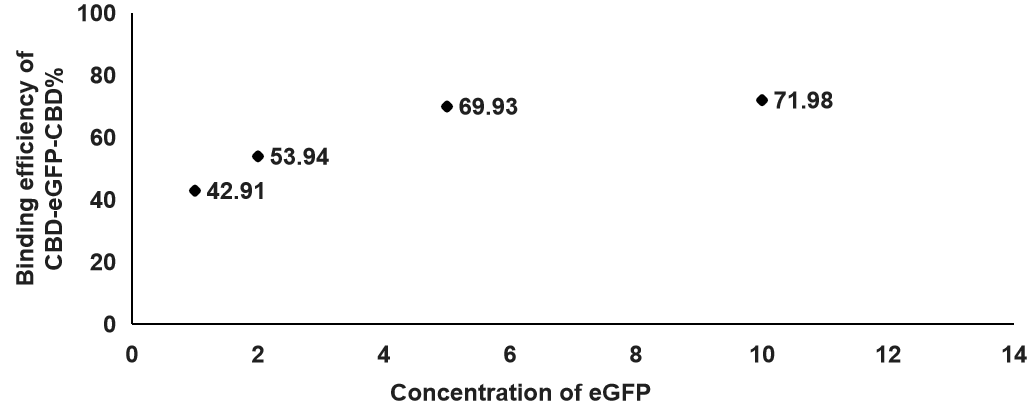
2.2After we determined which concentration of cellulose is best for binding, we also tested the optimal eGFP concentration for the binding efficiency between CBD-SA and cellulose. Additionally, the concentration of cellulose in this experiment is at 0.2%.
According to this figure, we found out that the binding efficiency was the highest when the concentration of eGFP was at 10, which means the binding efficiency was increasing when the concentration of eGFP was higher. That will be the concentration we use in the following experiments.
| None |