Part:BBa_K3791012
gRNA Erythromycin construct
This construct corresponds to the full DNA sequence that allows for the transcription of the guide RNA targeting erythromycin resistance. For that purpose, in addition to the gRNA, it includes a promoter. Particularly, it is the T7 promoter, which used in pertinent competent cells is indirectly inducible by the addition of IPTG (as the T7 RNA-polymerase needs IPTG to be expressed) and therefore permits transcriptional modulation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
1. Usage and biology: reporter and fluorescence
This part is used inside BL21 competent cells, where we cotransform a plasmid containing the current part, and another containing the LbCas12a (BBa_K2927005) protein. To build our plasmids we used Golden Gate assembly, and to transform them into BL21 cells we carried out Electroporation. More details about the building process are available on our Biosensor library building page.
In order to determine if our sensors worked properly we had to first induce our plasmids expression using IPTG (100uM) and then lysate the cells for liberating our CRISPR-Cas construct. The method used for that was enzymatic lysis (using a buffer without EDTA). After lysing our cells we proceed with the detection.
As context we must point out that for our CRISPR-Cas ensemble to be a detection system, what we did was to add a reporter to the media where the Cas may become active. The basis relies on LbCas12a’s special property of collateral trans-cleavage activity. That means that when it recognizes the specific target sequence determined by the designed gRNA with which it is coupled (thus becoming active), it unspecifically cuts not only the target DNA but also any ssDNA present in the media [1]. In this way, whenever Cas12a is activated, fluorescence arises because the reporter is cut and releases the fluorescent molecule. All this is step-by-step indicated in Figure 1.
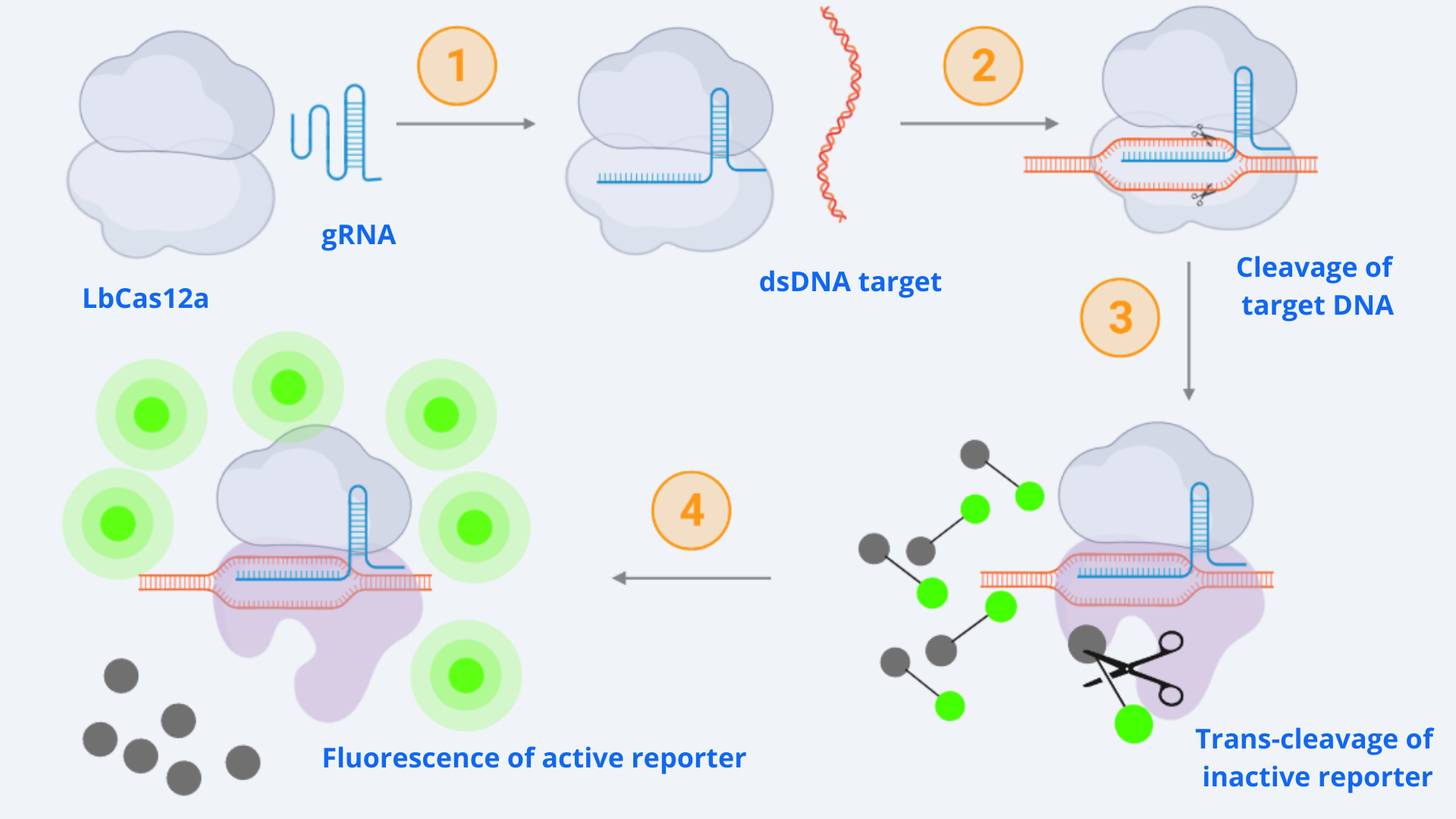
Figure 1 Description: General schematic of Cas12-gRNA-reporter functioning containing the main enzymatic reactions. Cas12 binds to the gRNA, this complex gets activated by recognizing the specific target and cis-cleavage activity starts by cutting this dsDNA target. After that, Cas12 trans-cleavage collateral activity cuts any nonspecific DNA fragment, cleaving the reporter. The reporter contains a quencher, which inhibits the fluorophore, and when cutted, luminescence comes out.
2. Characterization: model
CRISPR-Cas12 enzyme kinetics model was created by ARIA with the aim of understanding the dynamics and limitations of using the CRISPR-Cas technology for detection purposes. The model is extensively explained in our more complete composite part: BBa_K3791021, since the experimental parameters and results used for comparing with simulations were the ones performed with this part.
3. Characterization: Experimental results
3.1. Testing conditions
The fluorescence emitted by our system cannot be seen by naked eye, it needs to be checked with special light. Moreover, if we desire not only to determine its presence but also quantify it, we use special machinery as a Plate Reader. In our case, quantifying the amount of fluorescence resulting from our reactions was especially important as since we performed the detections in lysed bacterial cells there were lots of DNases, apart from Cas, cutting the reporter. The specifications for the configuration of the Plate Reader which are specified in Table 1 were designed by us, adapting the wavelength values to the ones specified in the reporter user manual. More details about measurement conditions are available in our Results measurement page.
| Parameters | Value |
| Temperature | 37ºC / Room temperature |
| Duration | 2h |
| Use kinetic interval | Measure every 5 minutes |
| Excitation | 530 nm |
| Emission | 560 nm |
| Number of flashes | 20 |
| Measure from | Bottom |
| Gain | Optimal (100 RFA) + Use Gain Regulation |
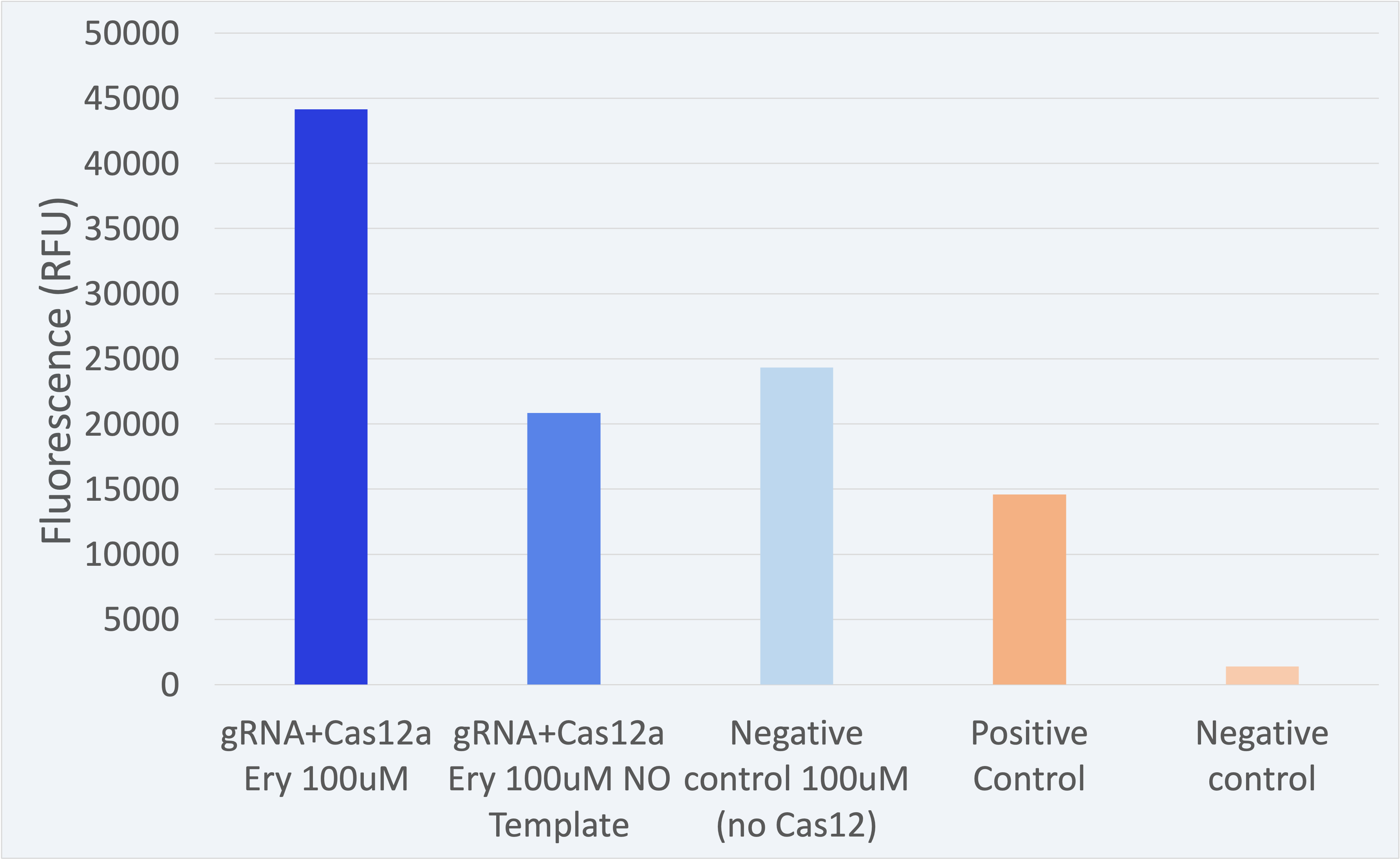
3.2. Best detection results
All experiments have been conducted under controlled conditions to respect procedural consistency and thus achieve the highest reproducibility of the results. For the results shown below, three biological replicates were used to ensure trustability.
The fluorescence results for this composite part + LbCas12a (BBa_K2927005) are shown in Figure 2.
The results shown above as successful, a distinguishable difference between the positives and negatives can be seen. Therefore, the results confirm our engineered biological system serves as a biosensor and accomplish the purpose for which it was created.
References
[1] Nguyen, L. T., Smith, B. M., & Jain, P. K. (2020). Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nature Communications, 11(1). https://doi.org/10.1038/s41467-020-18615-1//function/crispr/grna/construct
| None |