Part:BBa_K3724005
Lactoferrin Aptamer
Aptamer for Lactoferrin This part selectively detects Lactoferrin, with a detection limit of 1 ± 0.12 nM as determined in solution binding assays[1]. Lactoferrin, a protein involved in innate defense, has been indicated to have a major anti-inflammatory role in maintaining homeostasis[2,3]. Since organ failure is a common outcome of severe sepsis infection, lactoferrin has been shown to play a crucial role in indicating an overactive immune response[3].
Usage and Biology
Aptamers can be used to detect biomarkers in various body fluids, or to treat certain diseases by selectively inhibiting specific targets and therefore their signaling cascade[4]. Their function is similar to antibodies, but they are much easier to synthesize as they can be made in vitro via PCR.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Modelling
(Section added by iGEM 2022 team IISER_Mohali)
2D folding of Aptamer
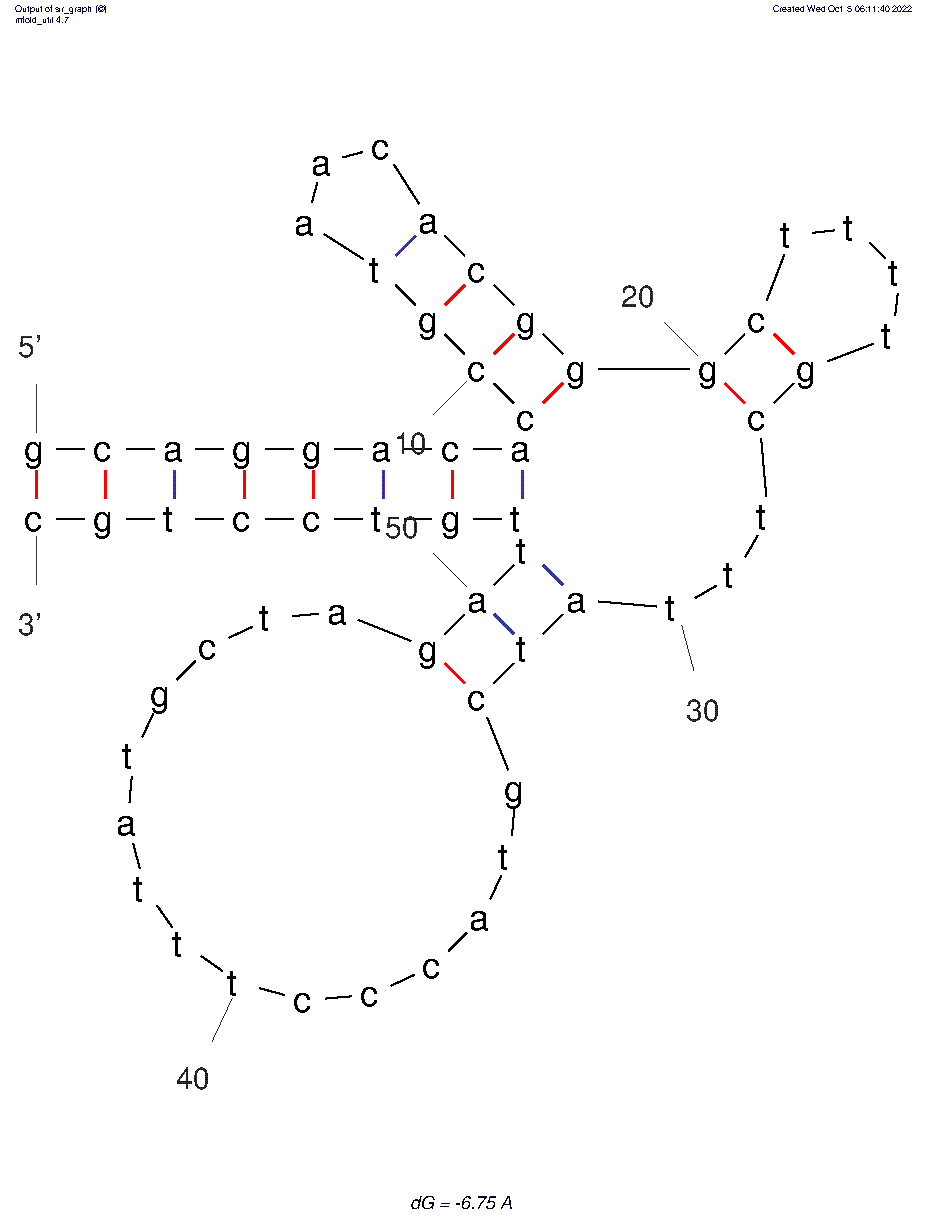
The 2D folded structure of the aptamer was generated using mFOLD (UNAFOLD web server). The input was the DNA aptamer sequence and default parameters 25°C, 1.0 M Na+ and 0.0 M Mg++, linear DNA, oligomer correction, 5 percent suboptimality. 3 structures were generated with structure 1 dG = -6.75 kcal/mol, structure 2 dG = -5.92 kcal/mol and structure 3 dG = -5.88 kcal/mol. Model-Fig-1 shows the least energy (most stable) structure 1.
3D structure of Aptamer
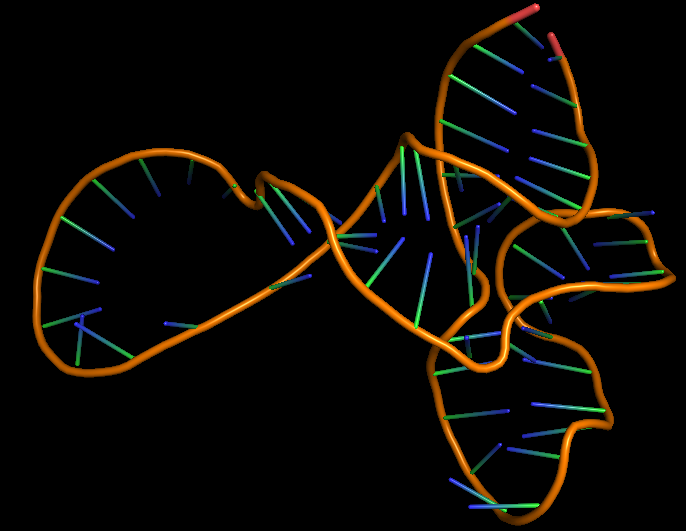
The 3D folded structure of the aptamer was generated using Xiao Lab 3dRNA/DNA - An RNA and DNA tertiary structure prediction method (http://biophy.hust.edu.cn/new/3dRNA/create). The input was the DNA aptamer sequence, the 2D structure in Vienna format, obtained from mFOLD and parameters molecule type DNA, Procedure Best, # of Predictions 5. No changes were made in Advanced options. Model-Fig-2 shows the most stable folded 3D structure.
Molecular Docking
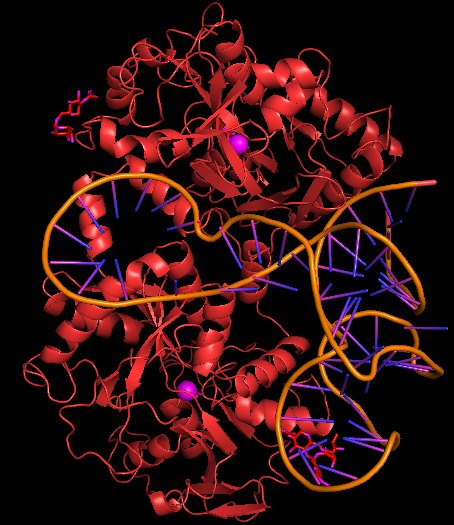
Human diferric lactoferrin, PDB ID 1LF6 was chosen at the protein that aptamer binds to. HDOCK server (http://hdock.phys.hust.edu.cn/) was used for docking aptamer and 1LF6. Help for using HDOCK server: http://hdock.phys.hust.edu.cn/help.php#metric. The Receptor 3D aptamer structure and the Ligand 1LF6 protein in pdb format were uploaded. Rest of the parameters were kept default.
Model-Table-1 shows the top 10 best docked models. According to HDOCK, a Confidence Score above 0.7 means that the two molecules would be very likely to bind. As all of the top 10 models have high Confidence Score of >8.0, IL-6 and the aptamer are likely to bind very well. Model-Fig-3 shows model_1 which is the most likely docked configuration (lowest Docking Score and highest Confidence Score).
Characterization
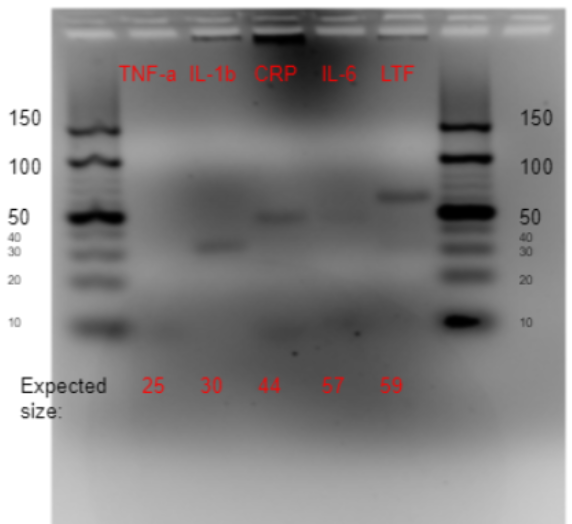
The aptamer was stored in E. coli inside a pUC-IDT Golden Gate vector with ampicillin resistance. This plasmid DNA was used as template for asymmetric PCR. The first PCR conditions (annealing gradient temperature 58-68°C and forward : reverse primer ratio of 20:1) showed that annealing temperatures around 60°C worked best (Figure 1, Table 1).
After going through multiple different conditions, the optimized PCR conditions were 20 cycles of amplification, 2 mM MgCl2 and 60°C annealing temperature. Figure 2 shows how the aptamer ran on 5% agarose gel, which confirms that it was successfully synthesized at the expected length.
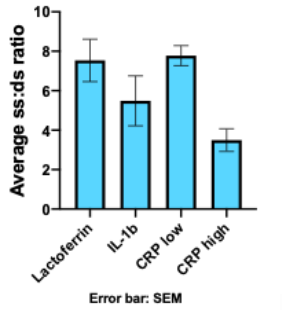
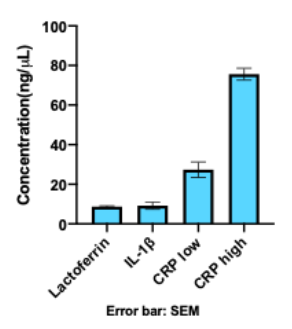
To determine the ratio of ssDNA : dsDNA that was synthesized, we used an assay that uses two different dyes, one with high affinity for ssDNA and one with high affinity for dsDNA, and the fluorescence is measured using a BioTek plate reader (Quantifluor assay). After performing six different reactions with these conditions, we obtained an average concentration of 8.68 ng/μL (Figure 4) and a ssDNA:dsDNA ratio of roughly 7:1 (Figure 3).
Aptamer attachment to reduced graphene oxide (rGO) and incorporation into electrode
The purpose of our aptamer biobrick is to show that it can be attached to rGO via π-π stacking, as well as detect the target biomarker when rGO is attached to the electrode and sweat flows through. For this biobrick, we were able to show that the aptamer attached to rGO through potentiostat measurements, and that it produces a change in resistance at different biomarker concentrations.
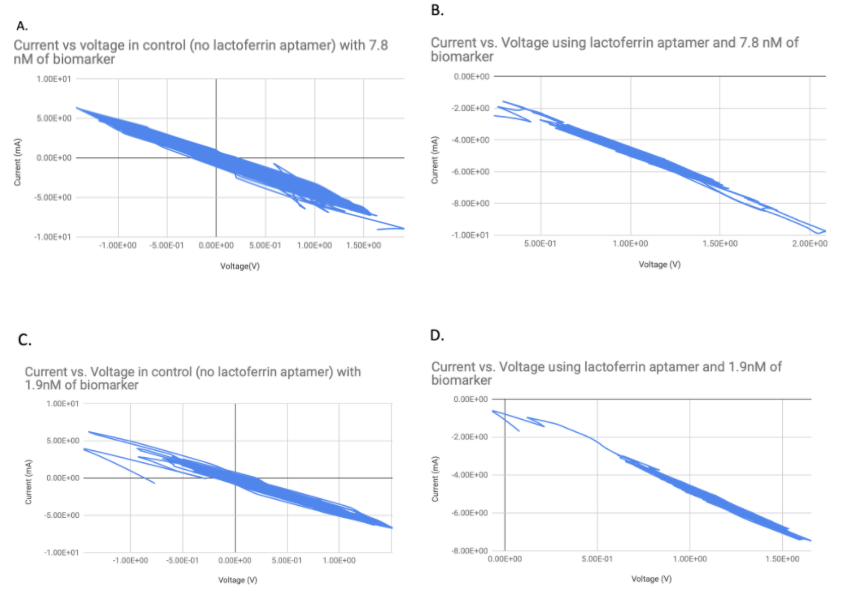
We performed the attachment in solution by mixing 10 mL of 4 g/L rGO with 2 mL 10 nM aptamer solution. As a control, we also attached 2 mL of just the buffer in which the DNA was. After 2 hours incubation at room temperature with end-over-end rotation, we gravity filtered to wash unbound DNA, and then we drop-casted the rGO-aptamer or rGO-buffer solutions onto screen printed electrodes. To determine if the lactoferrin aptamer successfully binds its target biomarker when attached to the rGO sheet, we tested the electrode modified with rGO + lactoferrin aptamer at different lactoferrin concentrations and monitored the change in resistance, via cyclic voltammetry.
As seen in Figure 5, we observed a shift of the peak current between the control (rGO, Figure 5 A and C) and experimental condition (rGO and aptamer, Figure 5 B and D) at all concentrations of biomarker, and, as an example, we showed the results of cyclic voltammetry for lactoferrin concentrations of 1.9 nM and 7.8 nM. We observed that the peak current for the control was about 6 mA at biomarker concentrations of 1.9 nM and 7.8 nM (Fig. 3A and 3C). We observed a shift in peak current between the control (rGO) and experimental condition (rGO and aptamer). The peak current on electrodes covered with rGO and lactoferrin aptamer was -2 mA and -1 mA for 7.8nM and 1.9nM respectively (Fig. 3B and 3D). The same trend was observed when the biomarker concentration of 1.9 nM was used. Thus, the peak current is higher in only buffer condition, which shows that there is an electrical change when the aptamer is attached to the rGO sheet.
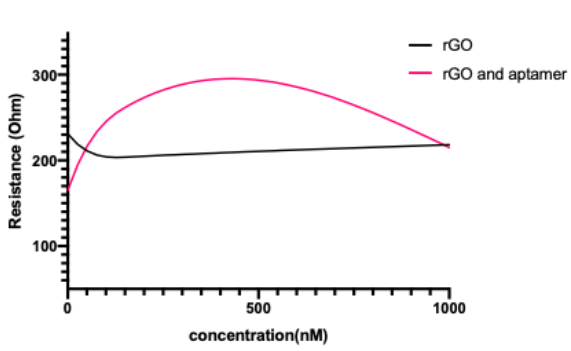
To analyze if there is a trend in electrical measurement with changing concentration, we used the peak current and inferred potential at which this current was observed to calculate the resistance change at each biomarker concentration condition. Our data shows significantly increased resistance when the aptamer is present as compared to the control with just buffer (Figure 6). The control was just a straight line, as the resistance values were around the same value for all of the biomarker concentrations. This data showed that there was not any non-specific binding and that binding occurred only in the presence of an aptamer attached to the electrode. The shift of the resistance in rGO + aptamer from the resistance in just rGO and its general increase indicates that the binding of biomarkers and aptamers can be measured using our potentiostat. We were able to detect lactoferrin in the 50 nM - 400 nM concentration range, which has clinical relevance[5].
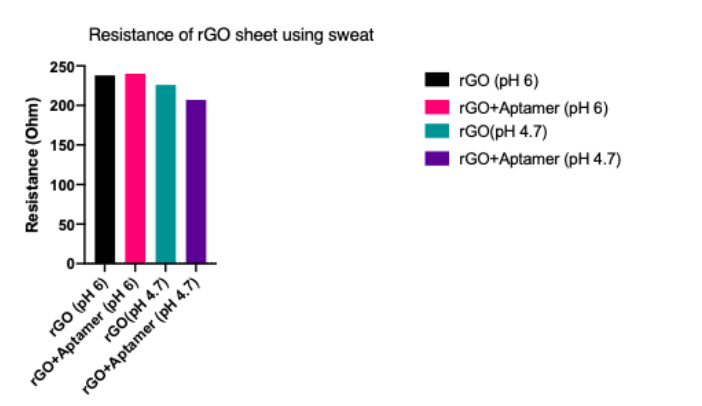
As an additional negative control, we tested the change in resistance when sweat at different pH was added to the electrode (modified with rGO + aptamer or just rGO). As seen in Figure 7, there is no change in resistance when only sweat is added, which confirms that the change observed previously is not due to some other molecule present in the sweat buffer, but rather due to the aptamer-biomarker interaction.
References
[1] Fang Yu, Hui Li, Wei Sun, Yaju Zhao, Danke Xu, Fuchu He, Selection of aptamers against Lactoferrin based on silver enhanced and fluorescence-activated cell sorting, Talanta, Volume 193, 2019, Pages 110-117, ISSN 0039-9140, https://doi.org/10.1016/j.talanta.2018.09.063
[2] Sánchez, L., Calvo, M., & Brock, J. H. (1992). Biological role of lactoferrin. Archives of disease in childhood, 67(5), 657–661. https://doi.org/10.1136/adc.67.5.657
[3] Kruzel, M. L., Zimecki, M., & Actor, J. K. (2017). Lactoferrin in a Context of Inflammation-Induced Pathology. Frontiers in Immunology, 8, 1438. https://doi.org/10.3389/fimmu.2017.01438
[4] Ho, D.-R.; Chang, P.-J.; Lin, W.-Y.; Huang, Y.-C.; Lin, J.-H.; Huang, K.-T.; Chan, W.-N.; Chen, C.-S., Beneficial Effects of Inflammatory Cytokine-Targeting Aptamers in an Animal Model of Chronic Prostatitis. International journal of molecular sciences 2020, 21 (11), 3953.
[5] Barkhatova NA. [The use of plasma lactoferrin in the diagnosis of pyonecrotic infections of soft tissues and sepsis]. Klin Med (Mosk). 2008;86(10):36-8. Russian. PMID: 19069457.
[6] Mfold web server for nucleic acid folding and hybridization prediction.
Nucleic Acids Res. 31 (13), 3406-15, (2003)
[7]SantaLucia, Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics.
Proc. Natl. Acad. Sci. USA 95, 1460-1465.
[8]Yi Zhang, Jun Wang, Yi Xiao, 3dRNA: 3D structure prediction from linear to circular RNAs[J]. Journal of Molecular Biology, 2022. doi:10.1016/j.jmb.2022.167452
[9] Yan Y, Tao H, He J, Huang S-Y.* The HDOCK server for integrated protein-protein docking. Nature Protocols, 2020; doi: https://doi.org/10.1038/s41596-020-0312-x.
[10] Yan Y, Zhang D, Zhou P, Li B, Huang S-Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365-W373.
| None |