Part:BBa_K3724001
IL-6 aptamer
Aptamer for Interleukin-6 (IL-6)
This part selectively detects IL-6, with a detection limit of 1.6 pg/mL, as measured on an AuNPs/pATP/pABA/GCE based aptasensor[1]. IL-6 is a pro-inflammatory cytokine, secreted by immune cells such as macrophages and helper T cells. It plays an important role in the innate immune response and thus shows elevated levels in sepsis, cancer and other immune disorders[2,3].
Usage and Biology
Aptamers can be used to detect biomarkers in various body fluids, or to treat certain diseases by selectively inhibiting specific targets and therefore their signaling cascade[4]. Their function is similar to antibodies, but they are much easier to synthesize as they can be made in vitro via PCR.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Modelling
(Section added by iGEM 2022 team IISER_Mohali)
2D folding of Aptamer
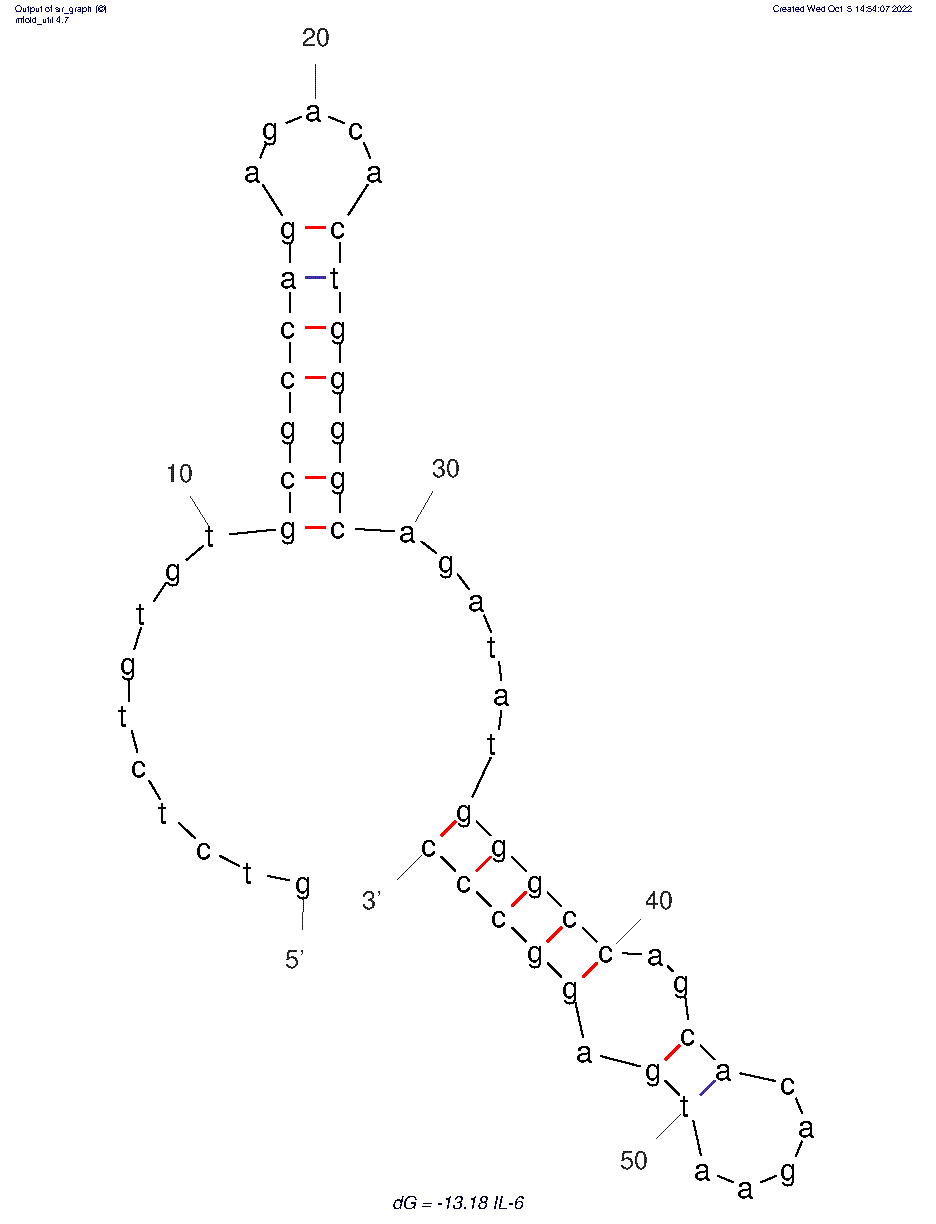
The 2D folded structure of the aptamer was generated using mFOLD (UNAFOLD web server). The input was the DNA aptamer sequence and default parameters 37°C, 1.0M Na+ and 0.0M Mg++, linear DNA, oligomer correction, 5 percent suboptimality. Rest of the parameters were also kept unchanged. Only one structure was generated with dG = -13.18 kcal/mol. Model-Fig-1 shows the 2D structure of the aptamer.
3D structure of Aptamer
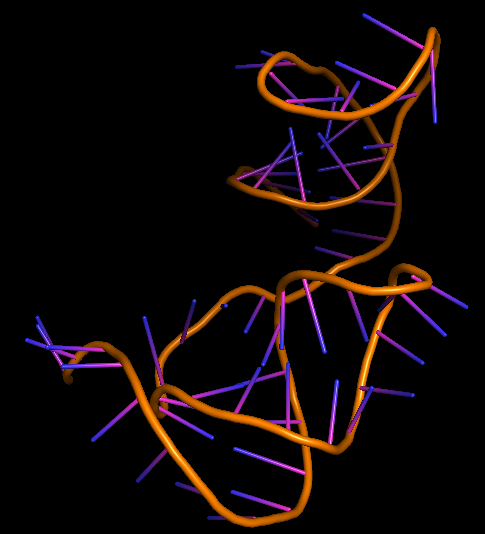
The 3D folded structure of the aptamer was generated using Xiao Lab 3dRNA/DNA - An RNA and DNA tertiary structure prediction method (http://biophy.hust.edu.cn/new/3dRNA/create). The input was the DNA aptamer sequence, the 2D structure in Vienna format, obtained from mFOLD and parameters molecule type DNA, Procedure Best, # of Predictions 5. No changes were made in Advanced options. Model-Fig-2 shows the most stable folded 3D structure.
Molecular Docking
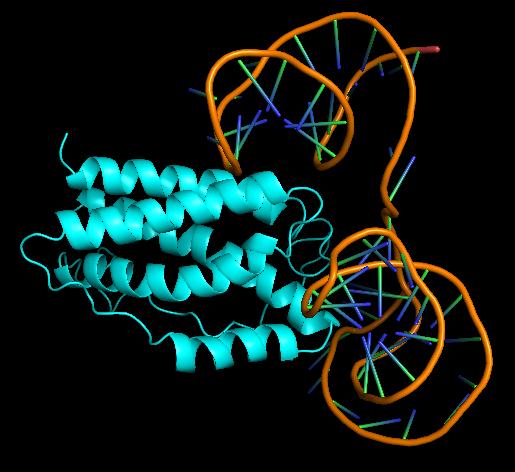
Human Interleukin-6, PDB ID 1IL6 was chosen at the protein that aptamer binds to. HDOCK server (http://hdock.phys.hust.edu.cn/) was used for docking aptamer and IL-6. Help for using HDOCK server: http://hdock.phys.hust.edu.cn/help.php#metric. The Receptor 3D aptamer structure and the Ligand 1IL6 protein in pdb format were uploaded. Rest of the parameters were kept default.
Model-Table-1 shows the top 10 best docked models. According to HDOCK, a Confidence Score above 0.7 means that the two molecules would be very likely to bind. As all of the top 10 models have high Confidence Score of >8.0, IL-6 and the aptamer are likely to bind very well. Model-Fig-3 shows model_1 which is the most likely docked configuration (lowest Docking Score and highest Confidence Score).
Characterization
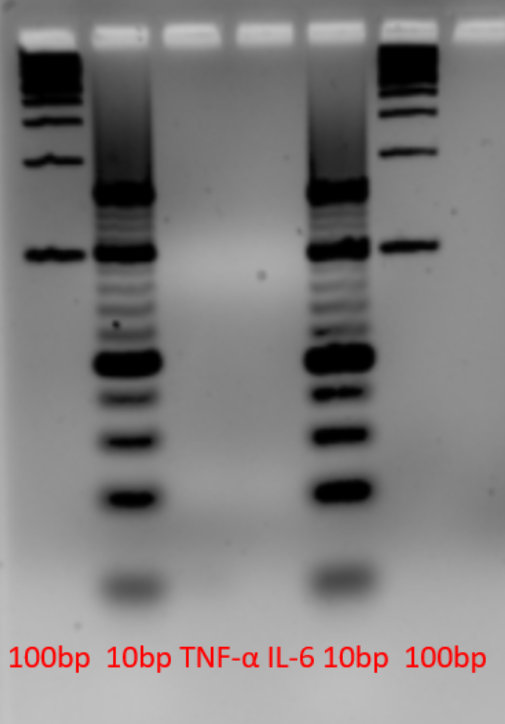
The aptamer was stored in E. coli inside a pUC-IDT Golden Gate vector with ampicillin resistance. This plasmid DNA was used as template for asymmetric PCR. From the first gradient PCR, we determined that higher annealing temperature gave more product for IL-6 (Figure 1).
The temperatures corresponding to each lane in Figure 1 are shown in Table 1.
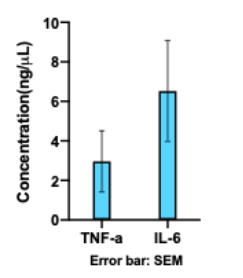
To optimize the PCR conditions, we ran multiple reactions with different number of cycles, Mg2+ concentration or forward:reverse primer ratio. Despite numerous tries to improve the single-stranded DNA concentration, we were not able to increase the concentration to more than 6 ng/μL, which was not sufficient for our future applications (Figure 2). Also, as seen in Figure 3, the aptamer was not visible on the gel. This could be because we stained with SYBR Gold which has a detection limit of 25 pg. Further improvements for this part could be changing the forward and reverse primers in asymmetric PCR, or visualizing the product on PAGE gel instead.
References
[1] Tertis, M.; Leva, P. I.; Bogdan, D.; Suciu, M.; Graur, F.; Cristea, C., Impedimetric aptasensor for the label-free and selective detection of Interleukin-6 for colorectal cancer screening. Biosensors and Bioelectronics 2019, 137, 123-132.
[2] Bozza, F. A.; Bozza Pt Fau - Castro Faria Neto, H. C.; Castro Faria Neto, H. C., Beyond sepsis pathophysiology with cytokines: what is their value as biomarkers for disease severity? (0074-0276 (Print)).
[3] Tanaka, T.; Narazaki, M.; Kishimoto, T., IL-6 in inflammation, immunity, and disease. (1943-0264 (Electronic)).
[4] Ho, D.-R.; Chang, P.-J.; Lin, W.-Y.; Huang, Y.-C.; Lin, J.-H.; Huang, K.-T.; Chan, W.-N.; Chen, C.-S., Beneficial Effects of Inflammatory Cytokine-Targeting Aptamers in an Animal Model of Chronic Prostatitis. International journal of molecular sciences 2020, 21 (11), 3953.
[5] Mfold web server for nucleic acid folding and hybridization prediction.
Nucleic Acids Res. 31 (13), 3406-15, (2003)
[6]SantaLucia, Jr (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics.
Proc. Natl. Acad. Sci. USA 95, 1460-1465.
[7]Yi Zhang, Jun Wang, Yi Xiao, 3dRNA: 3D structure prediction from linear to circular RNAs[J]. Journal of Molecular Biology, 2022. doi:10.1016/j.jmb.2022.167452
[8] Yan Y, Tao H, He J, Huang S-Y.* The HDOCK server for integrated protein-protein docking. Nature Protocols, 2020; doi: https://doi.org/10.1038/s41596-020-0312-x.
[9] Yan Y, Zhang D, Zhou P, Li B, Huang S-Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365-W373.
| None |