Part:BBa_K3686012
Chitinase from Xenorhabdus nematophila (strain 19061), complete CDs, codon-optimized.
Chitin, as one of the most abundant naturally occurring polymer, commonly found in fungal cell walls, the exoskeletons of invertebrates, and insect gut linings. In insects, the chitin layer provides extra support and shield the soft tissue against mechanical damages and toxins. Throughout evolution, many organisms have acquired ability to produce chitinolytic enzymes to digest chitin for nutrition. Since the catabolic reaction of chitin could be lethal for insect nymph and larvae, chitinases have great potency in the development of biological insecticide [1][2][3][4]. Besides, novel researches have shown enhanced toxicity of insecticides, such as Bt toxins, when mixed and use with chitinase [5].
[1]Mahmood, S., Kumar, M., Kumari, P., Mahapatro, G. K., Banerjee, N., & Sarin, N. B. (2020). Novel insecticidal chitinase from the insect pathogen Xenorhabdus nematophila. International Journal of Biological Macromolecules.
[2]Merzendorfer, H., & Zimoch, L. (2003). Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. Journal of Experimental Biology, 206(24), 4393-4412.
[3]Patil, R. S., Ghormade, V., & Deshpande, M. V. (2000). Chitinolytic enzymes: an exploration. Enzyme and microbial technology, 26(7), 473-483.
[4]Dahiya, N., Tewari, R., & Hoondal, G. S. (2006). Biotechnological aspects of chitinolytic enzymes: a review. Applied microbiology and biotechnology, 71(6), 773-782.
Considerations
pTac (T7 and LacO) regulated, His-tagged at C’ terminal for the ease of identification and purification.
Source
Isolated from insect pathogen Xenorhabdus nematophila (strain 19061), the gene ~1.9kbp in length coded for an endochitinase with molecular mass of 76kDa has been characterized by Kumar, M., Kant, S., Banerjee, N. and Sarin, N. B. and submitted to GenBank® under accession no: JN222361.
Expression with pTac system:
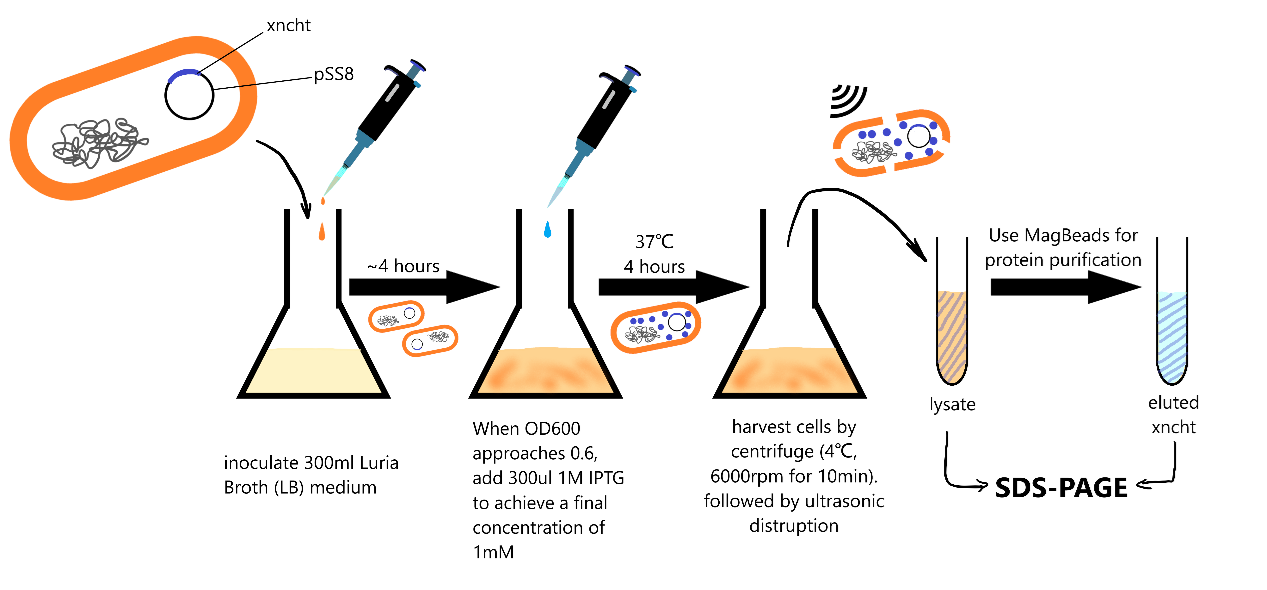
The sequence of chitinase gene form Xenorhabdus nematophila (abbreviated to xncht below) have been assembled with vector pET28a into plasmid pET28a-xncht, then transformed into E. coli (strain BL21) for expression:
Transformed strain was inoculated and cultured in 300ml LB medium at 37℃, when optical density (OD600) approaches 0.6, 1mM IPTG was added, the cells were harvested after 4 hours and disrupted to release the proteins in cytoplasm (detailed in our protocols and methods).
Analysis:
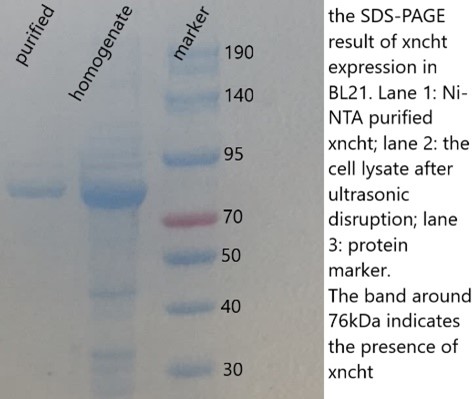
SDS-PAGE has been performed for both bacterial lysate (homogenate) and Ni-NTA beads resin (purified):
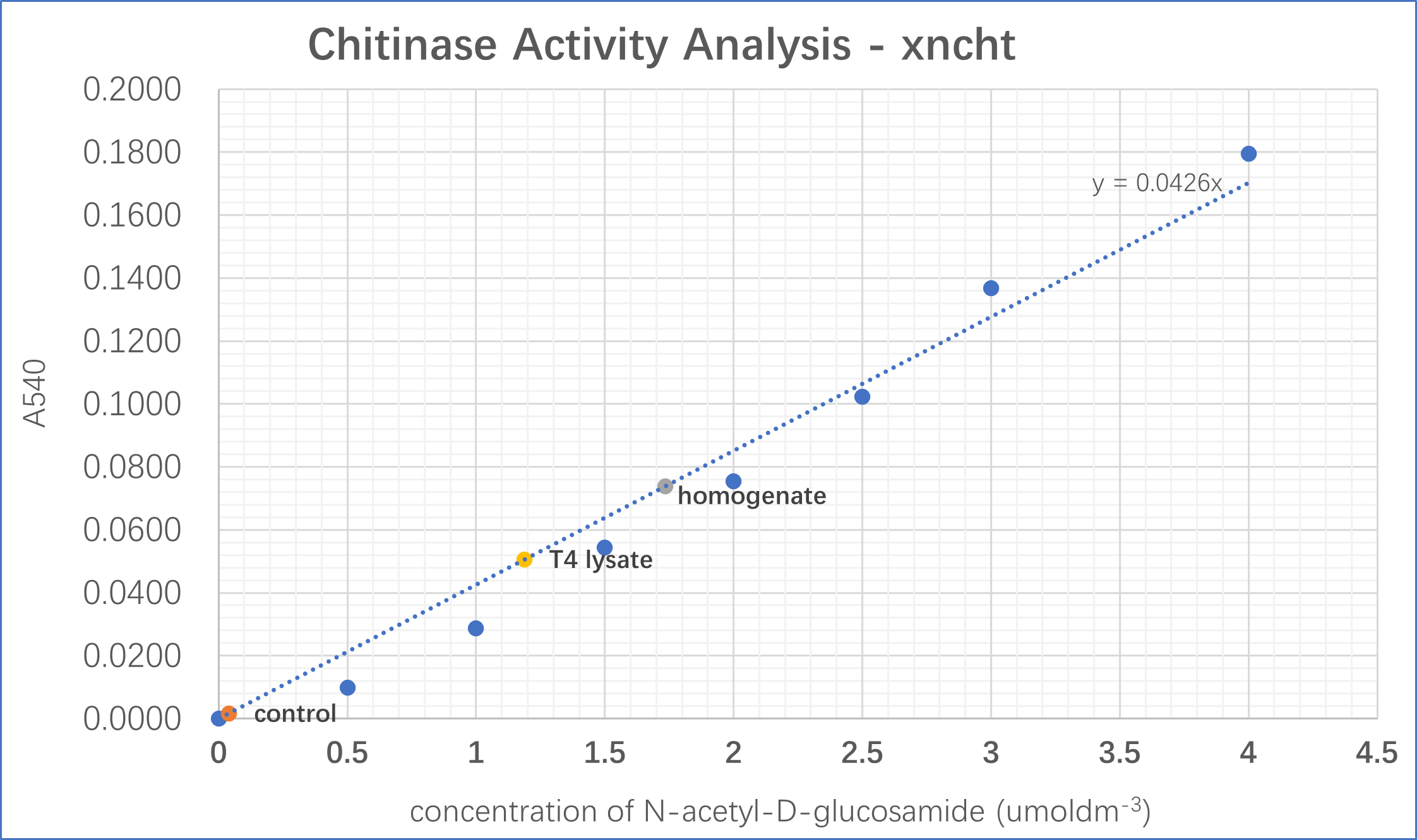
The chitinolytic activity of xncht has been investigated using the protocol suggested in Chitinase Assay Kit, (Solarbio, Beijing). Which defines the chitinolytic activity as the ability to hydrolyze chitin into its monomer N-acetyl-D-glucosamine (NAG) per hour. This monomer reacts with 3,5-dinitrosalicylic acid and form a red compound which absorbs light with 540nm wavelength . By comparing to the absorbance of standard solutions with different NAG concentrations, we could predict the number of monomers produced from catabolic digestion of chitin. Detailed in our protocols and methods .
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 756
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1599
Illegal AgeI site found at 1910 - 1000COMPATIBLE WITH RFC[1000]
| None |