Part:BBa_K3606029
P2 driven mcbABCD and PtetR driven mcbEFG
Background:
We used McbA-D to inhibit the growth of other bacteria in human intestine so as to enhance Nissle's competitiveness,as well as to reduce the risk of illness caused by some opportunistic pathogens. We introduced McbE-G to inhibit the growth of other bacteria in human intestine so as to enhance Nissle's competitiveness,as well as to reduce risk of illness caused by some opportunistic pathogens.At the same time, we put McbE-G under the control of pTetR in order to response to AHL-LuxR.
Design:
Here, we tried to improve the former antimicrobial peptide(mccb17) expressing system of 2019 Fudan BBa_K3245010. By dividing into the peptide expressing parts and the immunity parts, we wanted to manipulate their expression level with more efficiency.
Usage and Biology:
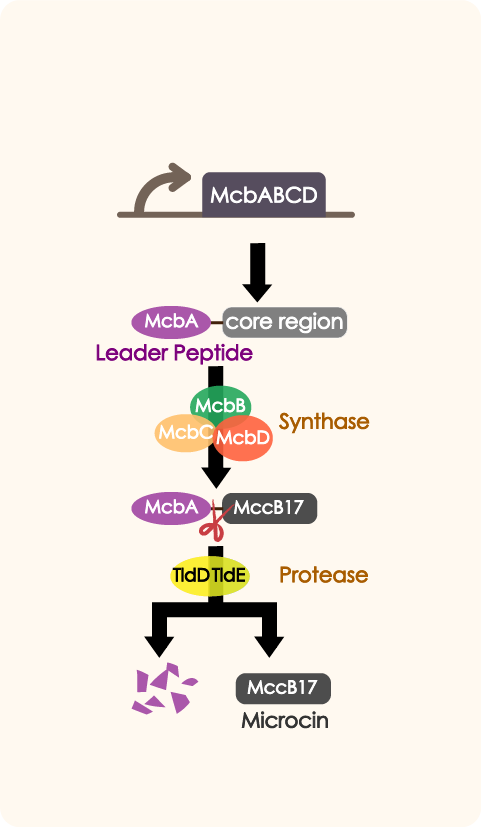
We introduced mcbABCD to our improved antibiotic expressing system to inhibit the growth of other bacteria in human intestine so as to enhance Nissle's competitiveness, as well as to reduce risk of illness caused by some opportunistic pathogens.Driven by a series of constitutive promoters with different strength which include P2, mcbABCD is expressed in E. coli in a controlled, reliable manner.
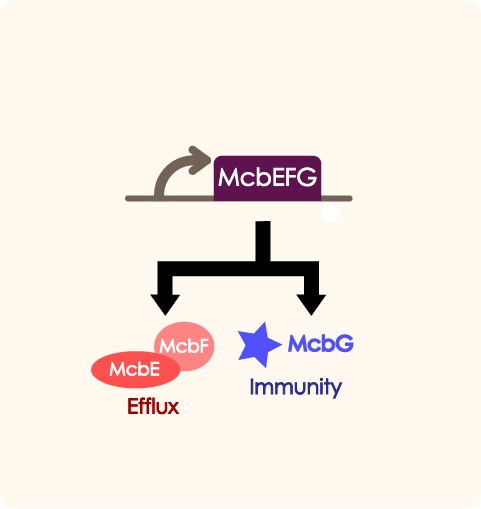
McbEFG works as an adjusting part in quorum sensing system. When the number of engineered bacteria is low, the part will help export antibiotic and provide self immunity in order to help proliferate. PtetR determines the expression of mcbEFG when the number of bacteria is at different level.
ATTENTION!The direction of the subparts shown in the following table is incorrect, our ptetR-McbEFG is a reverse sequence, and the correct look is as shown above! Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 2337
Illegal PstI site found at 2370 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 2337
Illegal PstI site found at 2370 - 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 2337
Illegal PstI site found at 2370 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 2337
Illegal PstI site found at 2370
Illegal NgoMIV site found at 1999
Illegal AgeI site found at 2171 - 1000COMPATIBLE WITH RFC[1000]
Methods:
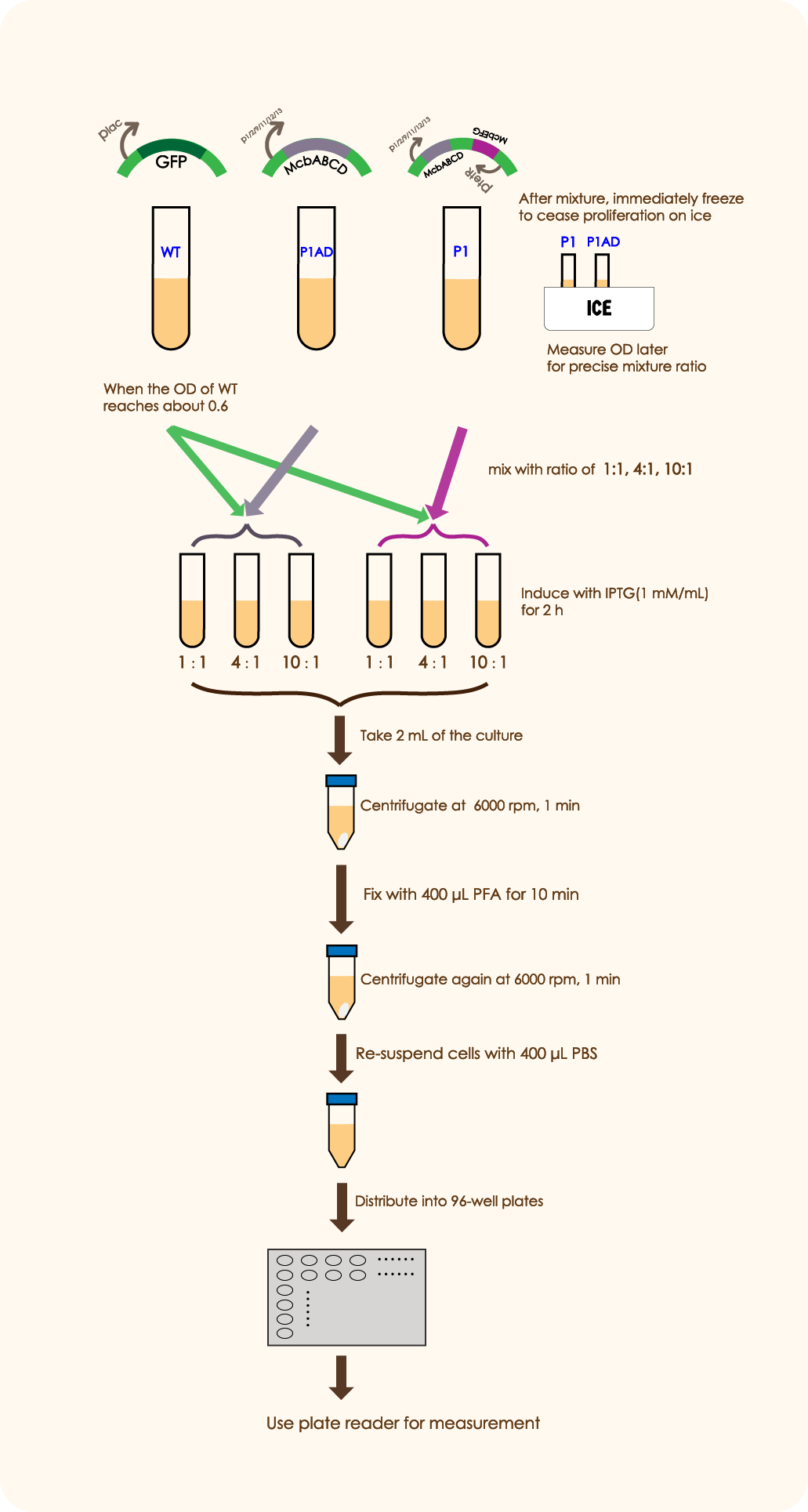
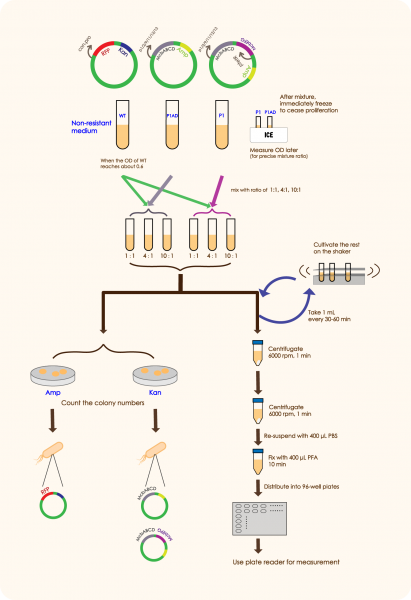
In order to prove that McbABCDEFG does have an effective antibacterial effect as well as McbEFG can protect the engineered bacteria themselves and help the secretion of antimicrobial peptides more effectively, we designed the following experiment:
We mixed WT E.coli (expressing GFP driven by plac) and E.coli with mcbABCD-mcbEFG-ptetR in different ratios (5:7 20:7 50:7), and measured the OD value of the bacteria two hours after adding an inducer, which can be used to reflect the antibacterial effect of antimicrobial peptides. We followed the same method as above to mix WT E. coli and E.coli with merely mcbABCD, induce and measure the OD value.
Results:
We constructed the plasmid and successfully expressed the system.
We could see an overall decrease of GFP expression in nearly all groups cocultured with P1 or P2AD than the control group which only contains WT, indicating that the antimicrobial peptide(MccB17) encoded by mcbABCD does have a negative effect on over microbial. In this case, the living status of WT cells and its function in creating GFP is strongly restricted by the toxic environment, proving that our part mcbABCD has worked successfully.
While inside each paired group, there clearly are a better inhibition effect in P1 group than P1AD group, as the MEFL/particle is much lower in the P1 group than the P1AD group when driven by certain promoters. This shows that the immunity function of mcbEFG is working successfully, as the E.coli expressing mcbEFG can help the engineered strain to survive with efflux exporting the toxic peptide, as well as better killing off other strains to gain survival advantage.
It is worth noting that when the mixing ratio of WT E. coli and McbABCDEFG-expressing bacteria is 50:7, the OD values of each group are very close to those of the control group (only induced WT E. coli). This shows that when the concentration of the engineered bacteria is very low, the antimicrobial peptides are difficult to exert antibacterial effect and the engineered bacteria have no competitive advantage.
We could see the population increase are not significant in total over time, yet WT grows rather fast at first in the P2AD group, indicating its lack of immunity part mcbEFG can lead to the death of engineered E.coli. more, thus shows less restrain on the WT cells. Yet, as mccb17 accumulates, a clear decrease can be observed when time extends in both P2 and P2AD, proving our antimicrobial peptide expressing system are working to help the engineered E.coli. gain better survival advantage.
Further Application:
For futher application, this part expresses antimicrobial peptide(mccb17) as well as providing immunity to create survival advantage for the engineered strain. These part are especially useful to be expressed in vivo because research has proved that it can also ease the inflammation in intestine by limiting the expansion of related pathogens and pathobionts.
References:
[1] Collin F, Maxwell A. The Microbial Toxin Microcin B17: Prospects for the Development of New Antibacterial Agents. J Mol Biol. 2019;431(18):3400–3426. doi:10.1016/j.jmb.2019.05.050
[2] S. Duquesne, D. Destoumieux-Garzón, J. Peduzzi, S. Rebuffat. Microcins, gene-encoded antibacterial peptides from enterobacteria
[3] Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcin
| None |