Part:BBa_K3257101
Cre Recombinase with SsrA(WVLAA)-aTc Inducible
For the best composite part, we would like to present you with our Cre recombinase together with degradation tag SsrA(WVLAA). Cre recombinase (BBa_K3257044 https://parts.igem.org/Part:BBa_K3257044) is responsible for the recombination process between the reverse-transcribed cDNA and the target gene, which is rather one of the most important processes in our system.
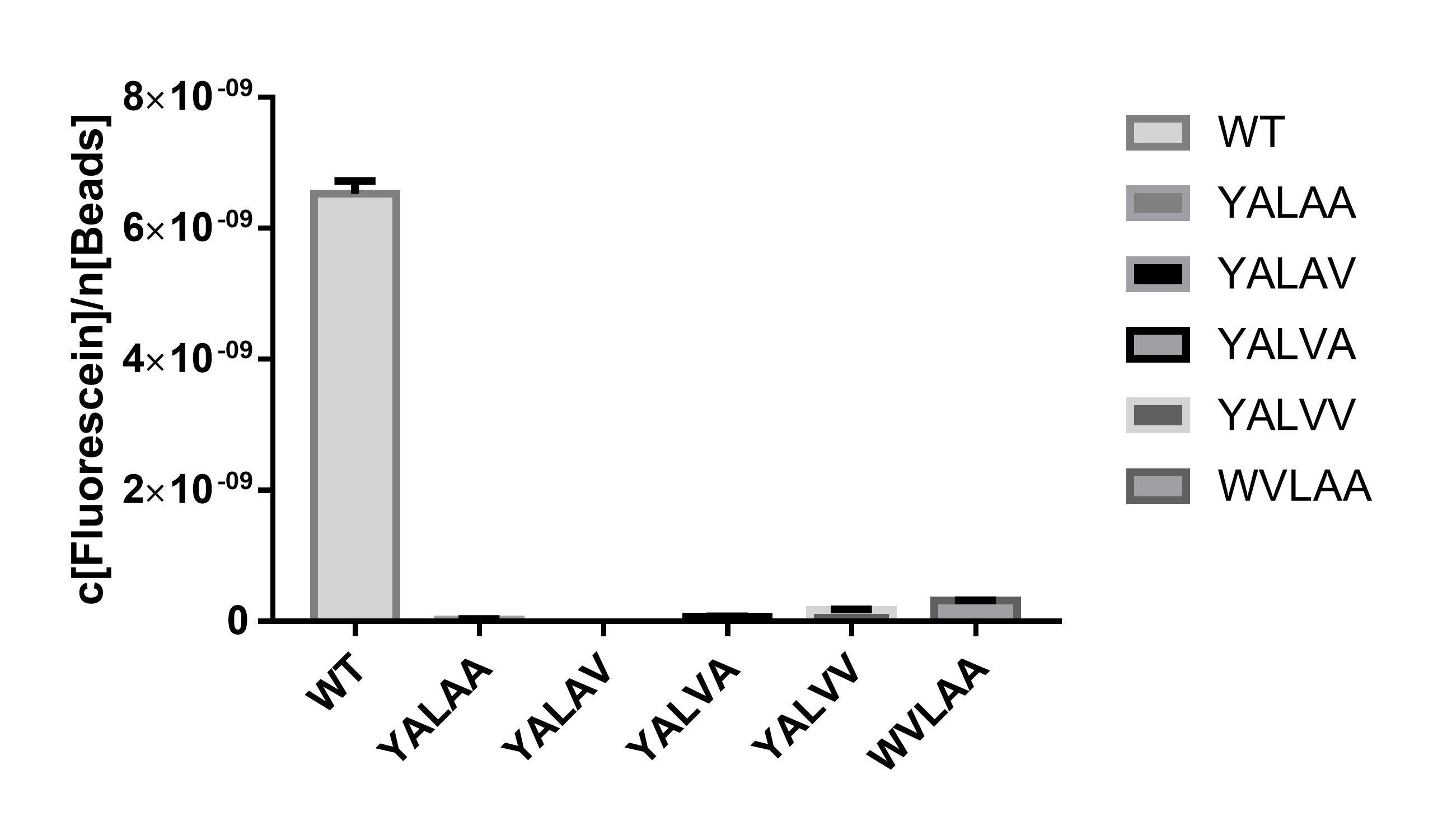
In order to deal with the leakage of uninduced Cre recombinase expression, which further establishing a more stringent control of the expression of Cre and agree to the modeling results that the concentration of Cre should be at a low level for the recombinant plasmids to accumulate, we have spent much effort in lowering the steady-state expression level of Cre. An important approach is to attach 5 different SsrA degradation tags after the CDS of Cre. Using EGFP (BBa_E0040 https://parts.igem.org/Part:BBa_E0040) as a reporter, we can see that the degradation rate of degradation tag with the amino acid residue sequence of AANDENWVLAA (BBa_K3257073 https://parts.igem.org/Part:BBa_K3257073) is moderate for Cre recombinase to reach a steady but relatively low level of expression (Figure 1).
Transformed into BL21 (DE3) with another plasmid containing lox sites and analyzed by colony PCR, we can see that DNA cleavage between identical-oriented loxP sites happens indicating the normal function of Cre recombinase(Figure 2).

Combining these two results, we show that this composite part is able to express Cre recombinase which functions well in an E.coli cell at a relatively low expression level, which is we need for new recombinants accumulating during the R-Evolution process.
In the future, iGEMers and synthetic biologists may use this part for homologous recombination with different degradation tags and promoters based on their own requirements.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 436
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 122
- 1000COMPATIBLE WITH RFC[1000]
| None |
