Part:BBa_K3078003
Bcam0581
Bcam0581 protein coding region. Bcam0581 is a functional homologue of rpfF. Bcam0581 is able to produce BDSF from the acyl carrier protein (ACP) thioester of 3-hydroxydodecanoic acid, a fatty acid synthesis intermediate.
1. Usage and Biology
Bcam0581, which has the activity of dehydratase and thioesterase, can produce the diffusible chemical BDSF through the intermediate substance in the fatty acid synthesis pathway of bacteria. BDSF can inhibit the phase transformation of C. albicans.
We characterised Bcam0581 by inserting it into pVE vector.
2. Characterization
2.1 Validation of pVE-Bcam0581 construction
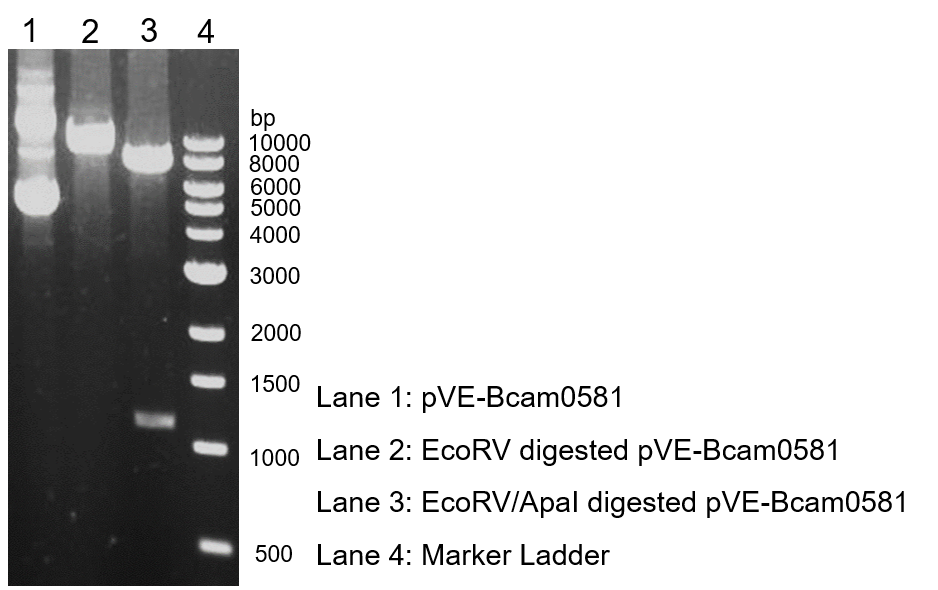
To verify the construction of pVE-Bcam0581 which we generated, the digestion by ApaI/EcoRV was performed by a standard protocol followed by agarose gel electrophoresis (Figure 1).
Figure 1. Digestion and electrophoresis of pVE-Bcam0581.
2.2 Expression of Bcam0581
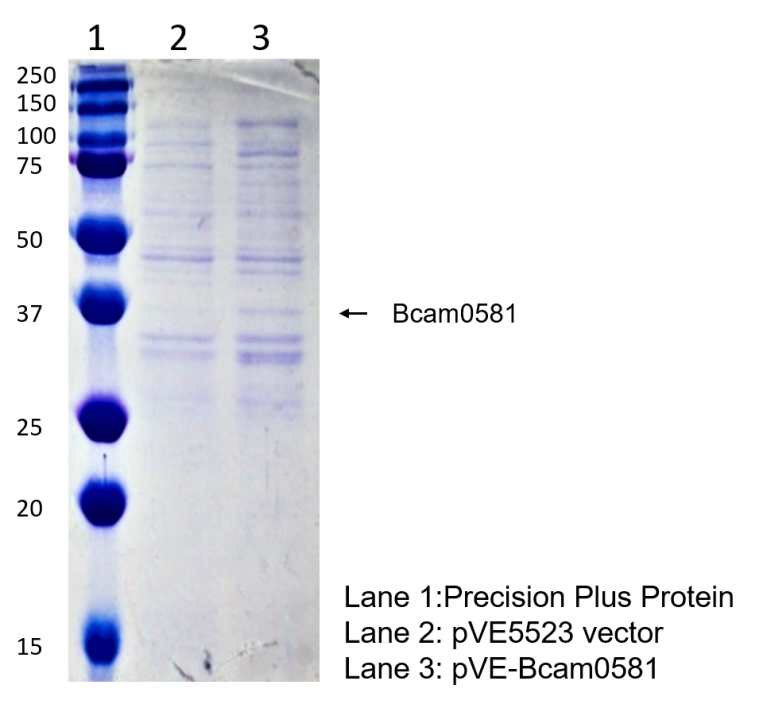
To detect the expression of Bcam0581 in E. coli, the constructs were transformed into BL21(DE3). Compared to the negative control, a distinct band at 37 kD was showed in pVE-Bcam0581(Figure 2).
Figure 2. Expression of Bcam0581. The bacteria were collected and ultrasonicated. The lysate was centrifuged and supernate was electrophoresed on the SDS-PAGE gel, followed by Coomassie brilliant blue staining.
2.3 The characterized of BDSF by HPLC
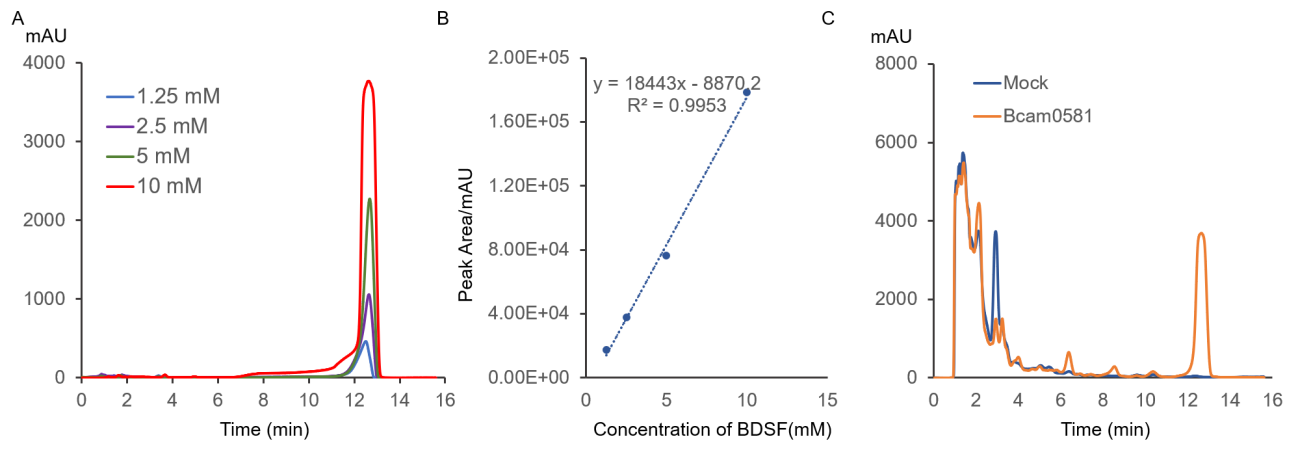
The relationship between the UV absorption peak area and BDSF concentration at 212 nm was measured with BDSF standards of different concentrations (Figure 3) to measure the concentration of BDSF generated by Bcam0581. The standard curve was established. pVE-Bcam0581 was transformed into E.coli BL21 (DE3) and BDSF concentrations in supernatant and negative control were measured.
Figure 3.A. HPLC analysis of BDSF standard with concentration of 1.25, 2.5, 5 or 10 mM showed that BDSF had the maximum absorption peak at 12.7 min. B. The standard curve was established for the absorption peak area at 12.7 min and BDSF standard concentration. The relationship between the peak area of BDSF standard HPLC results and BDSF in different concentrations: Peak area (mAU·s)=18443x(mM)-8870.2(mAU·s), R(sup)2(suped)=0.9953. C. Compared with the negative control (pVE5523 vector), the supernatant sample (pVE-Bcam0581) with 1000 times concentration had an obvious absorption peak with a peak area of 127352 mAU at the retention time of 12.7 min. By putting this result into the standard curve, the sample concentration was 7.39 mM, which meat that the BDSF concentration in the supernatant was 7.39 μM.
2.4 Candidacidal Effect of BDSF
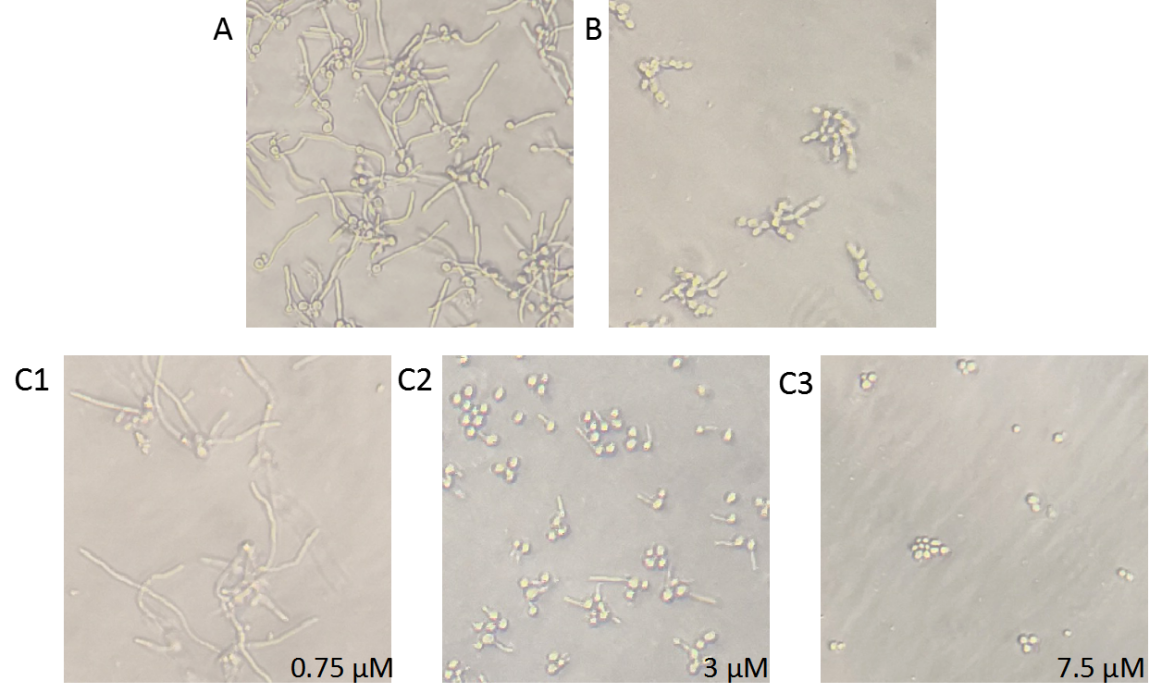
When BDSF standards and samples of different concentrations were added to the culture medium of C. albicans, 6-8 μM BDSF standards could completely inhibit the growth of hyphae. It can be seen from the experiments that the inhibition effect of supernatant samples on phase transformation is about the same as that of 6 μM BDSF standard, which can achieve the complete inhibition effect on hyhpae in theory (Figure 4).
Figure 4. BDSF inhibition of the phase transformation of C. albicans. C. albicans and the bacteria supernatant of control or Bcam0581 or BDSF standard were co-cultured in YPD medium supplementary with 10% serum in 96-well plate at 37℃ for 4 hours. After incubation, the hypha morphology was observed with inverted microscope. Three different pictures were selected from each well. A, mock control; B, bacteria supernatant of Bcam0581; C1~C3, different concentration of 0.75, 3 or 7.5 μM standard.
3. Conclusion
Our engineered bacteria successfully characterized Bcam0581. And it was proved that the BDSF produced by Bcam0581 was effective in inhibiting C. albicans phase transformation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 338
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 26
Illegal NgoMIV site found at 43
Illegal NgoMIV site found at 284 - 1000COMPATIBLE WITH RFC[1000]
| None |