Difference between revisions of "Part:BBa K3037002"
(→Overview) |
(→Overview) |
||
| Line 29: | Line 29: | ||
dCas9 was inserted into the pOCC97 vector [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000)] for transformation and expression in <span style="font-style: italic;">Escherichia coli (E. coli)</span>. | dCas9 was inserted into the pOCC97 vector [https://parts.igem.org/Part:BBa_K3037000 (BBa_K3037000)] for transformation and expression in <span style="font-style: italic;">Escherichia coli (E. coli)</span>. | ||
| − | There are many dCas9 BioBricks already available, however, all of them are optimized for the expression in mammalian cells. This is the very first dCas9 BioBrick that has been codon optimized for expression in <span style="font-style: italic;">E. coli</span>. Therefore, we are adding a new scope of in vitro applications of Cas9, which is normally used <i>in vivo</i>, to the iGEM community. | + | There are many dCas9 BioBricks already available, however, all of them are optimized for the expression in mammalian cells. This is the very first dCas9 BioBrick that has been codon optimized for expression in <span style="font-style: italic;">E. coli</span>. Therefore, we are adding a new scope of <i>in vitro</i> applications of Cas9, which is normally used <i>in vivo</i>, to the iGEM community. |
=== Biology === | === Biology === | ||
Revision as of 20:46, 21 October 2019
dead CRISPR Associated Protein (dCas9)
| dCas9 | |
|---|---|
| Function | Expression |
| Use in | Escherichia coli |
| RFC standard | Freiburg RFC25 standard |
| Backbone | pSB1C3 |
| Experimental Backbone | pOCC97 |
| Submitted by | Team: TU_Dresden 2019 |
Contents
Overview
The TU_Dresden 2019 team designed this BioBrick in order to make a fusion protein (BBa_K3037003) with dCas9 in accordance to the Freiburg RFC25 standard. (more information)
dCas9 was inserted into the pOCC97 vector (BBa_K3037000) for transformation and expression in Escherichia coli (E. coli).
There are many dCas9 BioBricks already available, however, all of them are optimized for the expression in mammalian cells. This is the very first dCas9 BioBrick that has been codon optimized for expression in E. coli. Therefore, we are adding a new scope of in vitro applications of Cas9, which is normally used in vivo, to the iGEM community.
Biology
dCas9 is a mutant derived from Cas9. Together with CRISPR, which stores sequences of viral infections, Cas9 forms a primitive antiviral defense system in bacteria. Coupled with a guideRNA, Cas9 has the ability to bind to a specific sequence of DNA and cut it. However, in contrast to its natural counterpart, dCas9 does not longer have the endonuclease activity. That means it is only binding to specific DNA sequences without cutting them. [1]
The targeted sequence is determined by the guideRNA bound to the dCas9 protein. The binding reaction was incubated at 37°C for one hour. We can put this system to use by providing guideRNAs to locate to any target DNA sequence with a high specificity.
Characterization
Overview
We performed the following characterization experiment to prove the DNA-binding ability of dCas9 via an Electrophoretic Mobility Shift Assay (EMSA) (Performed with BBa K3037005).
Experiments in Detail
1. Materials:
- 100 ng of PCR amplified sry gene
- 200 ng of dCas9-GFP
- 200 ng of guideRNA specifically targeting the amplified sry gene
- 1 x Reaction buffer - 20 mM HEPES buffer (pH 7.2)
- 100 mM NaCl
- 5 mM Mg{Cl{sub|2}}
- 0.1 mM EDTA
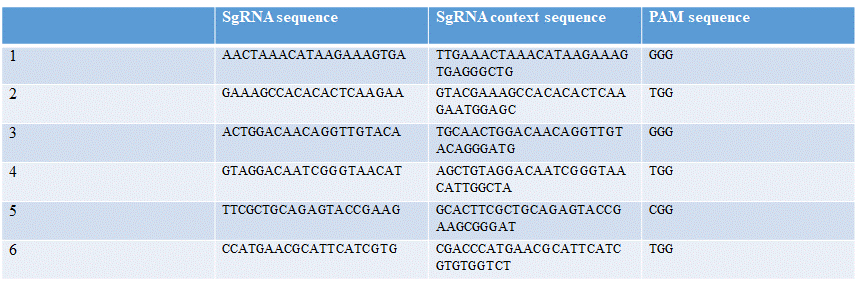
Six different guideRNAs were designed for targeting different regions of the sry gene, using the online tool "Benchling" and the FASTA sequence of the sry gene as seen in Table 1.
- AACTAAACATAAGAAAGTGA
- GAAAGCCACACACTCAAGAA
- ACTGGACAACAGGTTGTACA
- GTAGGACAATCGGGTAACAT
- TTCGCTGCAGAGTACCGAAG
- CCATGAACGCATTCATCGTG
2. Methods:
- We wanted to check if the overall efficiency of mobility shift increases when combinations of guideRNAs were used.
- GuideRNA, dCas9-GFP and sry gene were incubated in reaction buffer (respective amounts mentioned in the materials section) for 37 °C for 1 hour.
- Post incubation, they were mixed with a loading dye without SDS (20 % glycerol in Orange G dye) and loaded onto 4-20 % gradient acrylamide- TBE precast gel. Gel was run for 2 and 3 hours respectively, at 75V in 1 x TBE buffer.
- Gel was then stained using EtBR with 1:20000 dilution in 1x TBE for 10 minutes.
3. Results and Discussion
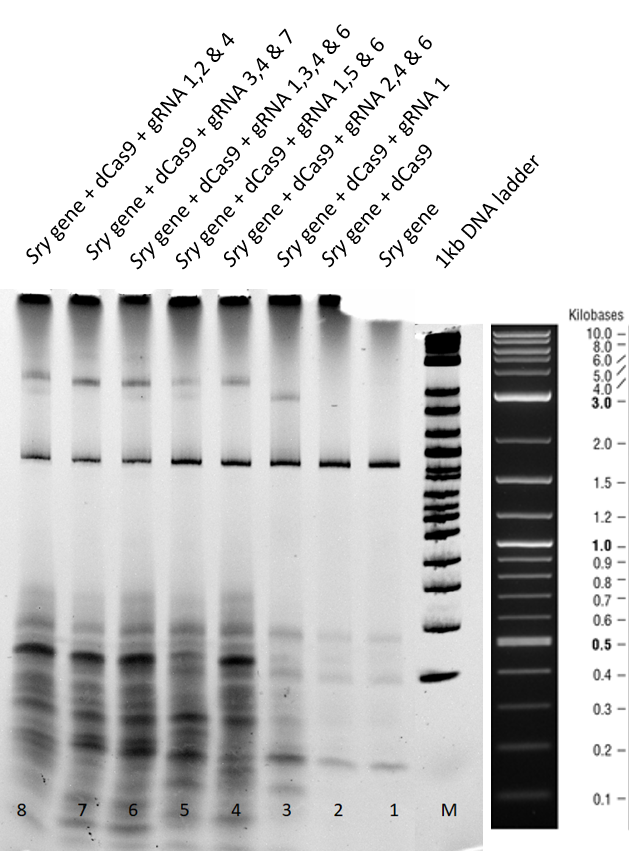
Gel run for 2 h:
A clear sry band at 800 bp is to be seen in lane 1 of Figure 1 when the sry gene is incubated is loaded seperately. When loaded with only dCas9 in lane 2, no shift is observed in the position of the band.
When guide RNA 1 was incubated with the dCas9 DNA reaction mix, we saw a shift in the mobility in lane 3. This is caused by the protein DNA interaction, where the binding is hindering the gene mobility.
In the lanes 5 to 9 different combinations of guideRNAs were deployed. In the lanes 7 and 8 we see the strongest mobility shift.
From the EMSA performed above, we conclude that our expressed dCas9-GFP protein is functional and is able to successfully bind to gene with the help of appropriate guideRNAs.
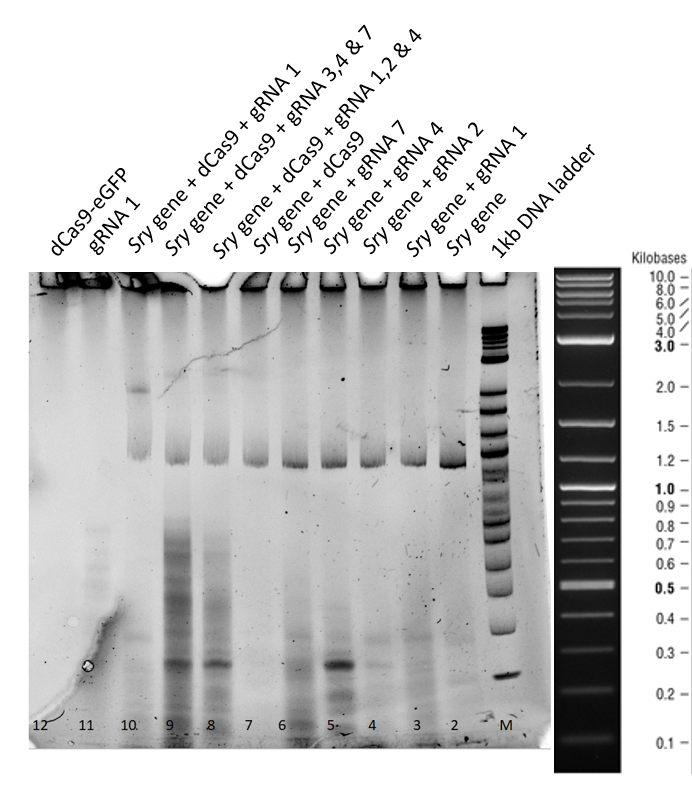
Gel Run for 3 h:
This second gel was run longer in order to get rid of all the secondary structures of the RNA formed.
From the Lanes 3 to 7 in Figure 2, no difference in the mobility of sry gene can be seen when only guideRNA is added to the reaction mix. In the Lanes 8, 9 and 10 a mobility shift of the gene can be observed and in lane 11, when only guideRNA was loaded, no bands were observed. In the Lane 12, dCas9 is in the stacking part of gel due to higher molecular weight.
4. Conclusions:
- We expressed functional dCas9, which is able to bind successfully to the sry gene with the help of our guideRNA.
- dCas9 on its own is unable to bind to the sry gene, suggesting that for binding guideRNA is required.
- GuideRNA on its own is unable to cause a mobility shift of the sry gene.
Sequence
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1096
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3375
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
In the middle of the coding sequence there was an EcoRI site. Therefore, we performed a site directed mutagenesis PCR with the following primers to erase the forbidden restriction site:
Primer 1: GATCGAATTCGCGGCCGCTTCTAGATAAGGAGGTCAAAAATGgccggcGATAAGAAATACTCAATAGGC
Primer 2: CATAATAAGGAATACGAAAAGTCAAG
Primer 3: CATAATAAGGAATACGAAAAGTCAAG
Primer 4: GATCTCTGCAGCGGCCGCTACTAGTATTAACCGGTGTCACCTCCTAGCTGACTCAAATC
The primers used to adapt this BioBrick to the Freiburg RFC25 standard were the following ones:
Prefix: GAATTCGCGGCCGCTTCTAGATAAGGAGGTCAAAAATGgccggc
Suffix: accggttaaTACTAGTAGCGGCCGCTGCAG
Find more information in here
References
[1] Whinn KS, Kaur G, Lewis JS, et al. (2019) Nuclease dead Cas9 is a programmable roadblock for DNA replication. Sci Rep. Sep 16;9(1):13292