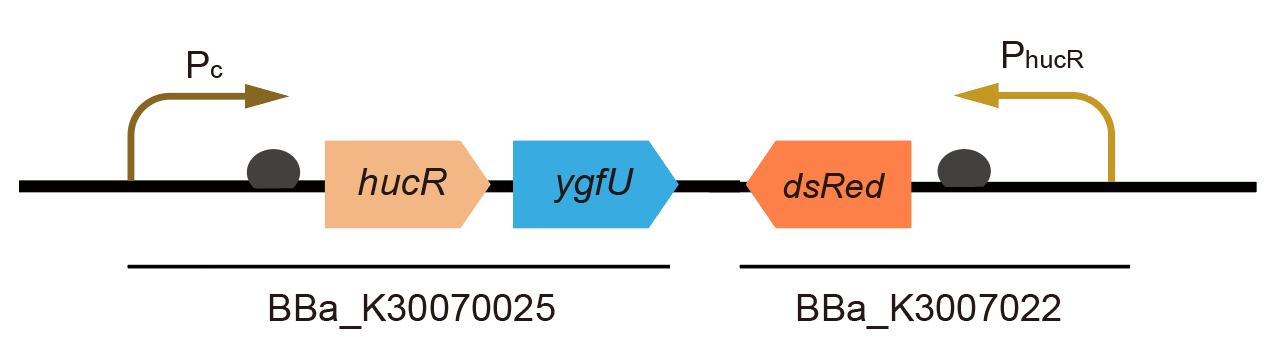
Part:BBa_K3007029
Expression device of dsRed under uric-acid-responsive regulatory system
By combining BBa_K3007022 and BBa_K3007025, we got a complete functional part which could transport and sense uric acid. When uric acid is absent, HucR can bind to PhucR, which suppresses dsRed expression. If uric acid presents in high concentration, HucR will release from PhucR and the expression of dsRed will recover from the inhibition [1]. In our design, by observing the expression of fluorescent proteins, we can know the concentration of uric acid in the environment.
Contribution
Group: QHFZ-China iGEM 2019
Author: Cheng Li
Design:
Improvement:
Previous: BBa_K2197300 (https://parts.igem.org/Part:BBa_K2197300)
New: BBa_K3007029 (https://parts.igem.org/Part:BBa_K3007029)
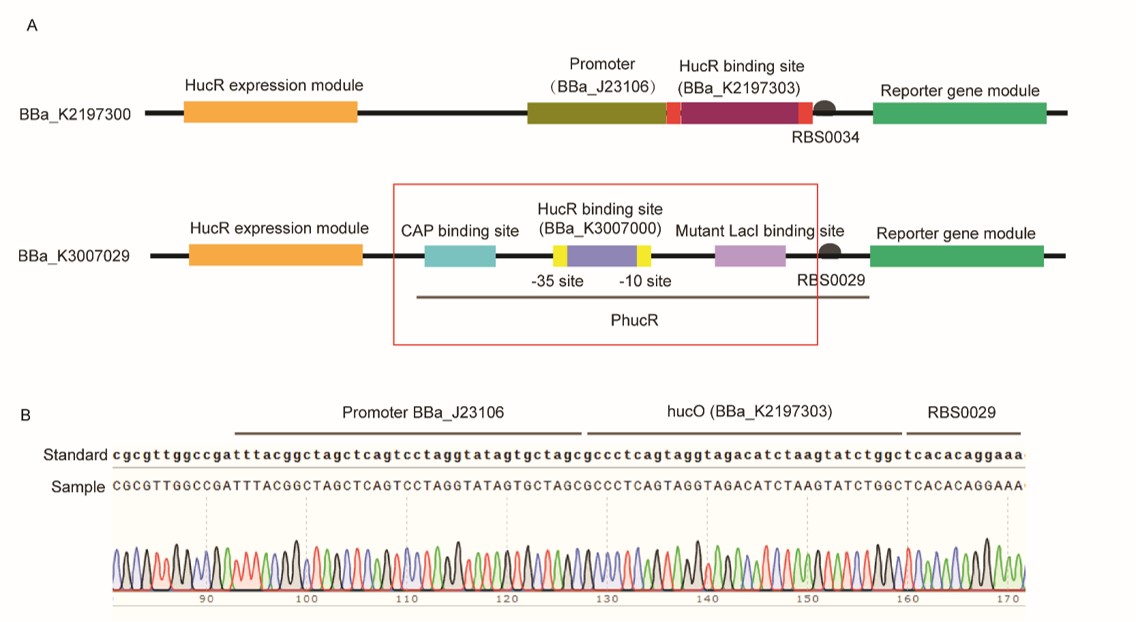
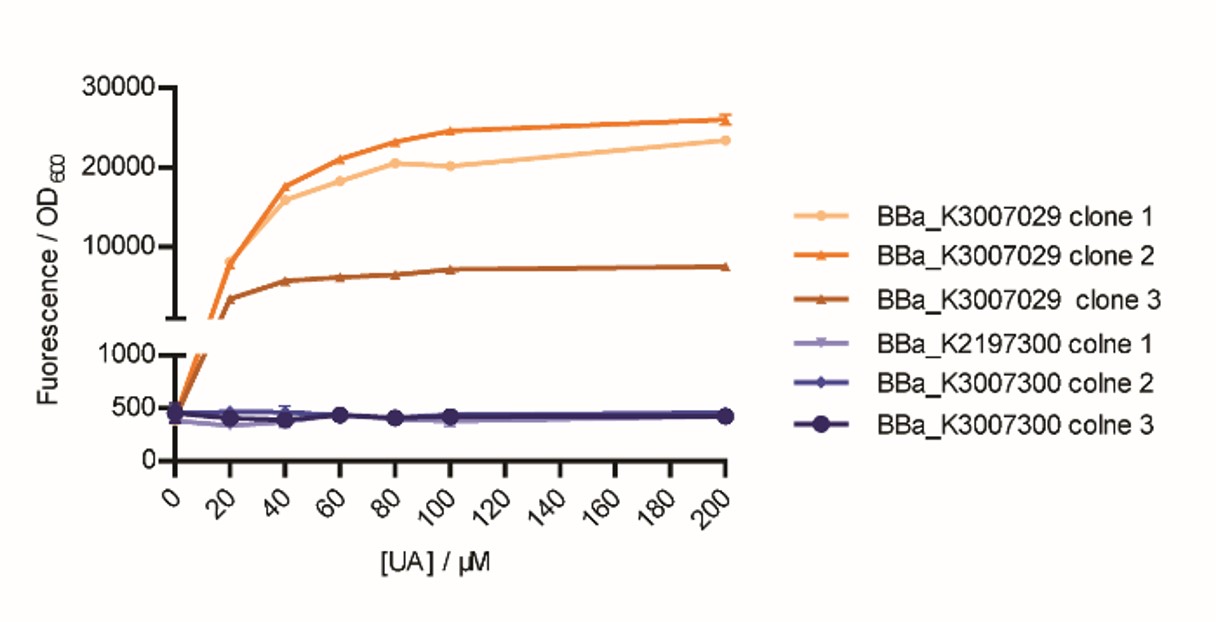
After we got our composite part BBa_K3007029 that can respond to uric acid (UA), we investigated whether the part worked better than a pre-existing part, BBa_K2197300. As the author team did not measured the part and declared the part they submitted could have something wrong, we decided to test the key difference of the two parts. Via analyzing, we found the HucR expression part is similar, while the most improvement of our part is using PhucR (BBa_K3007001) to control the expression of reporter gene and sense UA. Notably, considering that lac promoter is one of the most used promoter, we insert the hucO, which is the HucR binding sequence, between -35 and -10 regions of a modified lac promoter. However, in BBa_K2197300, the promoter BBa_J23106 and hucO sequence BBa_K2197303 connected end to end. As a result, we decided to construct a vector based on our vector by changing PhucR into BBa_J23106+ BBa_K2197303, to simulate the function of BBa_K2197300. As the 3’ end of lac promoter contains a RBS BBa_B0029 sequence, we retained the RBS sequence so that no more RBS is needed (Fig.1).
We chose three correct clones to do the experiment. We tested the response of the two parts to UA at the same time. The part that simulated the function of BBa_K2197300 could not respond to UA, while our BBa_K3007029 gave gradually enhanced fluorescence with the rise of UA concentration (Fig. 2).
Then we did more measurement and improvement of our BBa_K3007029. The detailed information has been shown below.
Documentation:
We used the process shown in Fig. 2 to test if the UA detection system works well.
Two clones with UA detection system with dsRed as a reporter were tested. The original gene circuit was able to response to UA in a range of 0 to 200 μM (Fig. 3A), and the clone 1 showed much better dynamics than the other (Fig. 3B). Time course experiments showed that the fluorescence intensity became quite strong at 4 to 6 hours after UA induction, and became stable at 10 to 12 hours (Fig. 3C). Even if we removed UA by replacing fresh LB medium, after 48 hours shaking, the fluorescence would be still notable (Fig. 3D) and there was not significant difference of dsRed fluorescence / OD600 between before and after UA removing (Fig. 3E). All the data meant our design could detect high UA concentration quickly and stably.

We also tried more conditions to test if this system could work well in different environment. In the range of pH 6.0 to 8.0, response of the gene circuit was relatively stable (Fig. 4A). However, the volume of the reaction system would influence the response to UA (Fig. 4B). A possible explain was the relative surface area of the liquid level changed and consequently the dissolved oxygen changed. This result meant the experiments for UA detection should be done at the same reaction system volume. In other experiments, 1 mL reaction volume was used.

Because the designed applications included to detect UA in blood or saliva sample, which contained serum, we tested if serum affects the detection efficiency. In view of safety, commercial fetal bovine serum (FBS) (EVERY GREEN, 11011-8611) was used here. The growth of the bacteria was obviously suppressed when the volume of FBS fraction was more than 1/1000 (Fig. 5A, 5B), which meant 1 μL fetal bovine serum was added to 1000 μL final reaction system. When the volume of FBS fraction was 1/1000, the UA detection efficacy was unaffected by serum (Fig. 5C).

We verified the designed system did response UA in different environments. However, during our human practices, some of interviewees worried about long responding time, which needed 4 to 6 hours after UA induction to express strong fluorescence intensity. In their opinions, users would not wait for a 4-hour reaction. And there was another feedback that the standard of hyperuricemia is ≥7 mg/dL (about 400 μM) UA for men and ≥6.0 mg/dL (about 350 μM) UA for women [2]. If we want to use our system to detect a clinical sample directly, the sample should be diluted to 1/1000 before start, which means the gene circuit is required to detect 400× 1/1000 = 0.4 μM UA as a threshold. In one word, we need a modification to shorten the responding time and increase the sensitivity of the UA detector.
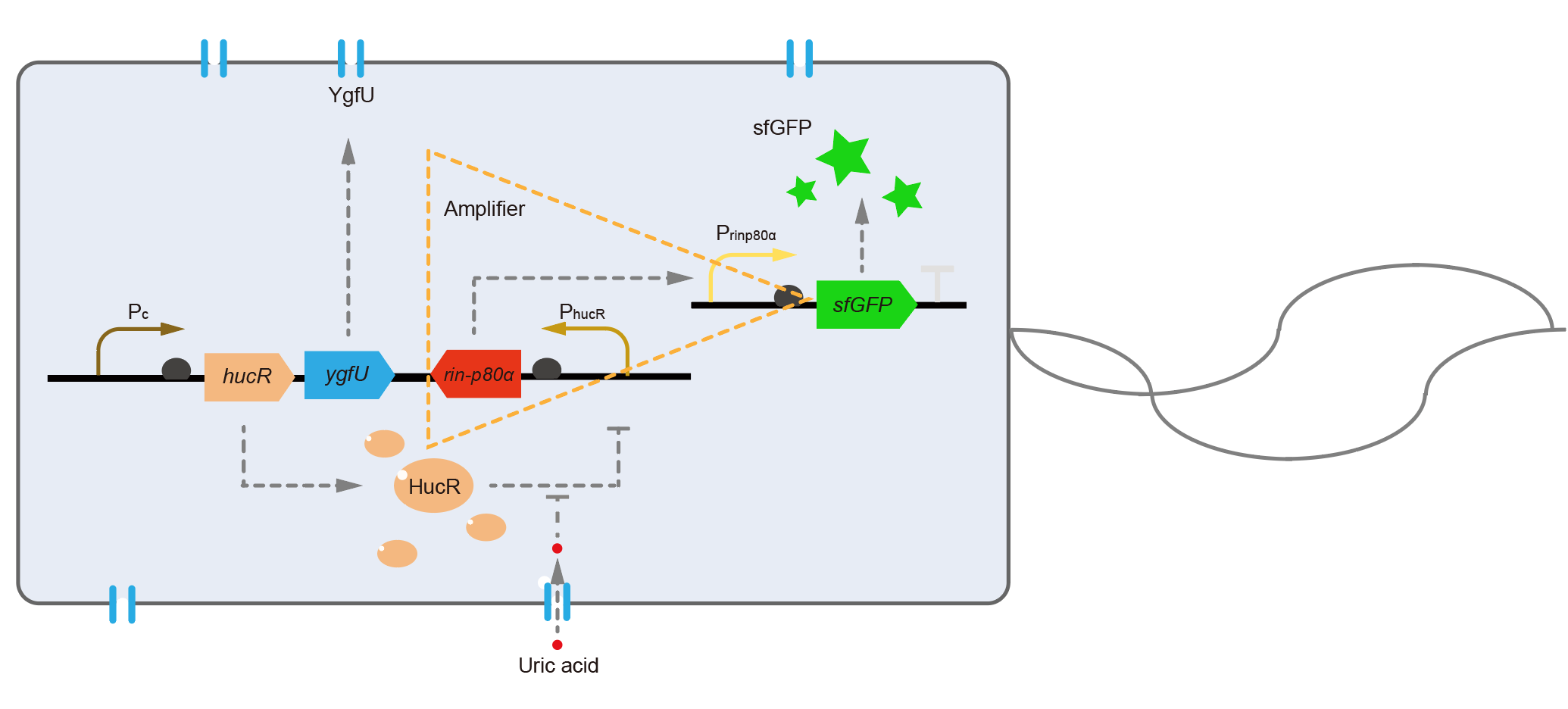
For the first question, we interviewed Dr. Xiaoyu Chen, a scientist who majors in biosensors. He suggested us that changing the fluorescent protein was a means to optimize the response speed. We blasted the sequence of dsRed we used in the database, and found the maturation half-time of dsRed is about 40 minutes in E. coli [3]. To shorten the maturation time, we decided to change dsRed to superfolder GFP, whose maturation half-time was only about 13 minutes [4]. To solve the second problem, we referenced a modular, cascaded signal amplifying methodology, which induces a module named amplifier, and it may increase sensitivity of the biosensor and boost the output expression [5]. We ordered two sequences of ultrasensitive phage activator RinA_p80α (from Staphylococcal aureus phage 80α) and a promoter PrinA_p80α. We introduced these new parts to design a new version of the UA detection system, called Version 2, shown in Fig. 6.
The processes in Version 2 were almost equal to the old version, except that the downstream of PhucR was RinA_p80α. This meant if UA presented, RinA_p80α would express and active transcription of sfGFP which was under control of PrinA_p80α. Theoretically, the new design would sense UA with much higher sensitivity than the old one. In the same time, the fluorescence production of Version 2 would get faster because that sfGFP had a shorter maturation time than dsRed in old version.
We tested the sfGFP production of Version 2 under different concentration of extracellular UA. The curve in Fig. 7A showed the fluorescence was saturated under only 15 μM UA induction, while the old version needed about 100 μM UA to get saturated (Fig. 3B). This result verified the Version 2 had higher sensitivity than the old one. To test if sfGFP could shorten the reaction time, we used the same construct only except reporter genes, called PRinA_p80α – sfGFP and PRinA_p80α – dsRed, respectively. After adding 20 μM UA into the reaction system, the curve of PRinA_p80α – sfGFP climbed much faster than PRinA_p80α – dsRed, which suggested our new design had a great induction performance, and fitted our predictions very well.
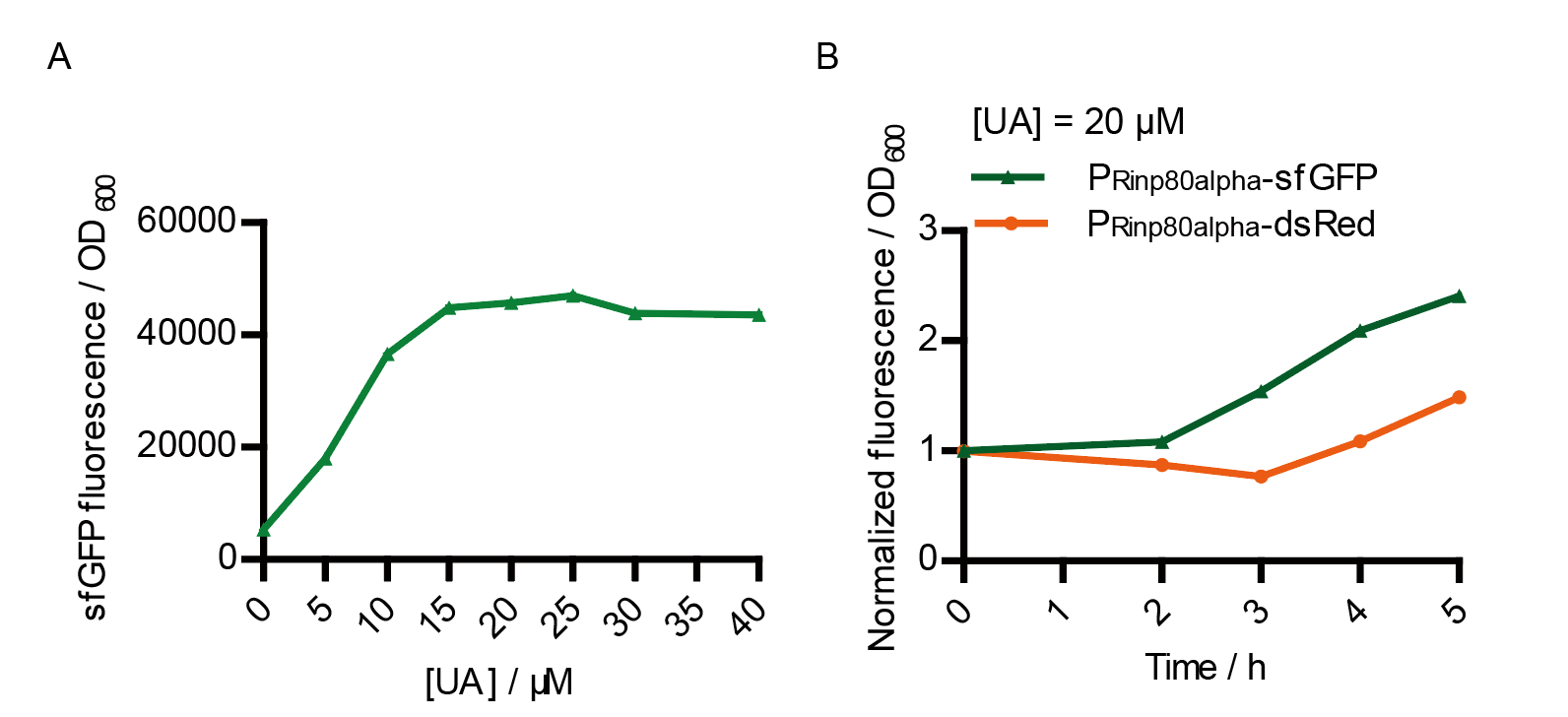
However, the induction time and sensitivity of Version 2 were still not reach the clinic required. In the future, we will optimize the gene circuit. We can change the RBS sequence of RinA_p80α and the RBS of sfGFP. We can also change the pSB1C3 plasmid which carries PrinA_p80α – sfGFP in Version 2, to plasmids with higher or lower copy number. Even, we can try to introduce more layers of amplifier to get high-gain amplification of output, which may make our system more sensitive.
References:
[1] Wilkinson, S. P., & Grove, A. (2004). HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. Journal of biological chemistry, 279(49), 51442-51450.
[2] de Oliveira, E. P., & Burini, R. C. (2012). High plasma uric acid concentration: causes and consequences. Diabetology & metabolic syndrome, 4(1), 12.
[3] Bevis, B. J., & Glick, B. S. (2002). Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nature biotechnology, 20(1), 83.
[4] Pédelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C., & Waldo, G. S. (2006). Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology, 24(1), 79.
[5] Wan, X., Volpetti, F., Petrova, E., French, C., Maerkl, S. J., & Wang, B. (2019). Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nature chemical biology, 15(5), 540.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 1399
Illegal EcoRI site found at 3216
Illegal XbaI site found at 3113
Illegal SpeI site found at 917
Illegal PstI site found at 140
Illegal PstI site found at 510
Illegal PstI site found at 3210 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1399
Illegal EcoRI site found at 3216
Illegal NheI site found at 1085
Illegal SpeI site found at 917
Illegal PstI site found at 140
Illegal PstI site found at 510
Illegal PstI site found at 3210 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1399
Illegal EcoRI site found at 3216
Illegal BglII site found at 852
Illegal BamHI site found at 158
Illegal BamHI site found at 2266
Illegal BamHI site found at 3130
Illegal BamHI site found at 3205 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 1399
Illegal EcoRI site found at 3216
Illegal XbaI site found at 3113
Illegal SpeI site found at 917
Illegal PstI site found at 140
Illegal PstI site found at 510
Illegal PstI site found at 3210 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 1399
Illegal EcoRI site found at 3216
Illegal XbaI site found at 3113
Illegal SpeI site found at 917
Illegal PstI site found at 140
Illegal PstI site found at 510
Illegal PstI site found at 3210
Illegal AgeI site found at 2084 - 1000COMPATIBLE WITH RFC[1000]
| None |