Difference between revisions of "Part:BBa K2965021"
| Line 19: | Line 19: | ||
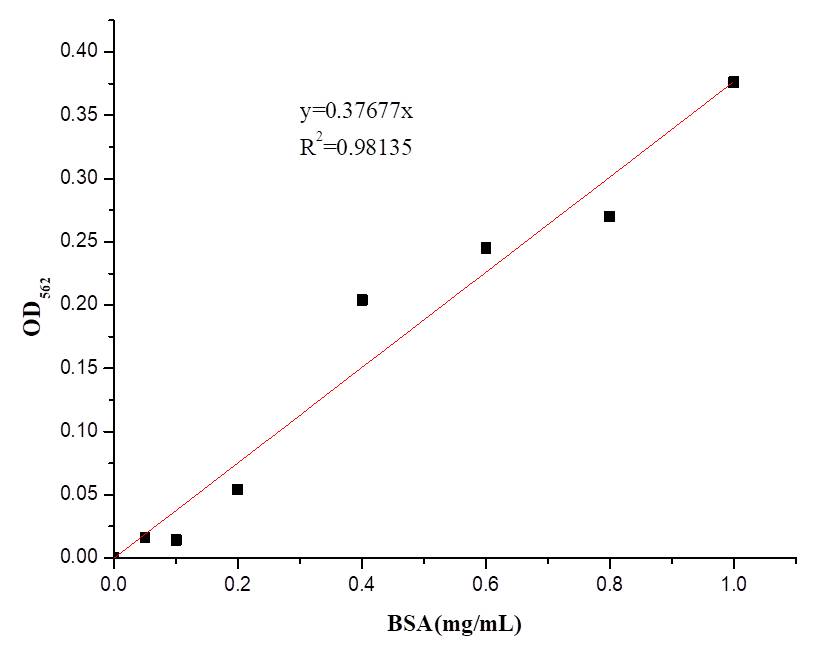
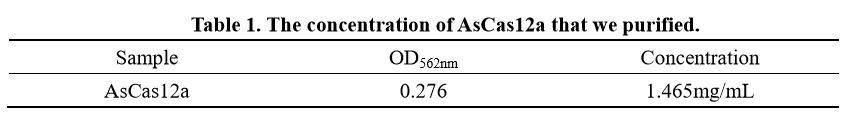
| − | In order to better characterise the AsCas12a expression and purification, we also determined the concentration of AsCas12a purified with BCA kit(Vazyme Biotech Co.,Ltd). | + | In order to better characterise the AsCas12a expression and purification, we also determined the concentration of AsCas12a purified with BCA kit(Vazyme Biotech Co.,Ltd) and the result is shown in Table 1. |
[[File:BBa K2965021-Figure 4. BSA standard curve..png|center|500px|thumb|'''Figure 4. The BSA standard curve.''']] | [[File:BBa K2965021-Figure 4. BSA standard curve..png|center|500px|thumb|'''Figure 4. The BSA standard curve.''']] | ||
[[File:BBa K2965021-Table 1. AsCas12a concentration..png|center|500px|thumb|]] | [[File:BBa K2965021-Table 1. AsCas12a concentration..png|center|500px|thumb|]] | ||
| − | Finally, we | + | Finally, we tested the activity of AsCas12a. We used a kind of 5bp single strand DNA with 5'-FAM and 3'-TAMRA as reporter in which the fluorescence of FAM can be quenched by TAMRA to display its DNA endonuclease activity. When AsCas12a is activated and cuts reporters, the cleavage of reporters will enable FAM to emit green fluorescence and neutralize the red fluorescence of TAMRA to be yellow. With same target DNA and crRNA, our extracted AsCas12a showed the ability to cut DNA compared with the control group and a significantly higher activity compared with the bought AsCas12a (shown as Figure 5.). |
[[File:BBa K2965021-Figure 5. AsCas12a activity assay..png|center|500px|thumb|''' Figure 5. AsCas12a activity assay.''']] | [[File:BBa K2965021-Figure 5. AsCas12a activity assay..png|center|500px|thumb|''' Figure 5. AsCas12a activity assay.''']] | ||
Revision as of 16:15, 21 October 2019
AsCas12a (AsCpf1)
AsCas12a (AsCpf1) comes from Acidaminococcus sp. BV3L6, it is a programmable DNA endonuclease guided by a single CRISPR RNA (crRNA). Targeting requires a crRNA complementary to the target site as well as a 5' TTTN protospacer adjacent motif (PAM) on the DNA strand opposite the target sequence. Once it recognizes its target, it will be activated and exhibits a “collateral effects” of promiscuous DNase activity.
Usage and Biology
AsCas12a RNPs bind to DNA and start to move along it by a hopping mechanism. When the AsCas12a RNPs encounter a potential target site containing a PAM sequence, they begin to initiate R-loop formation by forming base-pair hybrids between the crRNA and the target DNA strand. If sufficient matched nucleotides exist in the PAM-proximal region, the R-loop formation will be stable. After the stable R-loop conformation is formed, AsCas12a, via the activity of its RuvC domain, first cleaves the non-target DNA strand and then cleaves the opposite, target DNA strand, regardless of the presence of a PAM sequence. Finally, AsCas12a rapidly releases the PAM-distal DNA fragment but continues to hold the PAM-proximal portion of DNA[1].
Characterization
Result
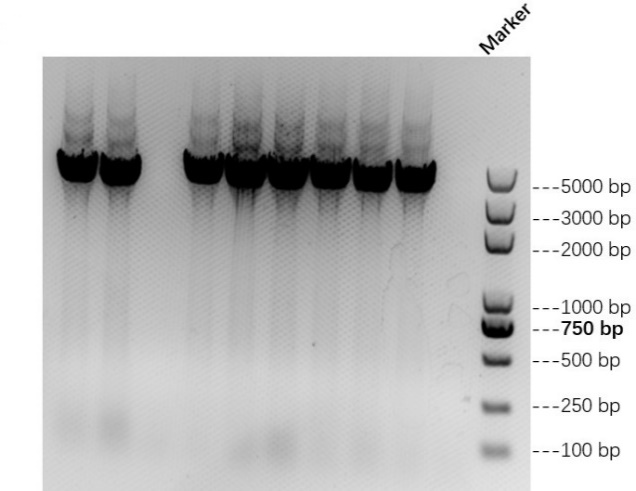
We linked AsCas12a fragment with His-tag and pET-28a(+) plasmid to construct the AsCas12a expression vector. Then we transfered these recombinant plasmids into E.coli BL21(DE3) and picked single colonies to carry out colony PCR and sequencing (by TSINGKE Biologocal Technology Institute) to ensure that we make successful transformation. The result of colony PCR is shown in Figure 1 below.
We induced AsCas12a's expression with IPTG and made purification using gravity-flow column with Ni-NTA Sefinose(TM) Resin. SDS-PAGE and Western Blot are carried out to test protein purification as shown in Figure 2 and Figure 3.
In order to better characterise the AsCas12a expression and purification, we also determined the concentration of AsCas12a purified with BCA kit(Vazyme Biotech Co.,Ltd) and the result is shown in Table 1.
Finally, we tested the activity of AsCas12a. We used a kind of 5bp single strand DNA with 5'-FAM and 3'-TAMRA as reporter in which the fluorescence of FAM can be quenched by TAMRA to display its DNA endonuclease activity. When AsCas12a is activated and cuts reporters, the cleavage of reporters will enable FAM to emit green fluorescence and neutralize the red fluorescence of TAMRA to be yellow. With same target DNA and crRNA, our extracted AsCas12a showed the ability to cut DNA compared with the control group and a significantly higher activity compared with the bought AsCas12a (shown as Figure 5.).
Reference
[1] Jeon Y, Choi YH, Jang Y, Yu J, Goo J, Lee G, et al. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nature Communications. 2018;9(1):2777-11.
[2] Mohanraju, P., Oost, J. v., Jinek, M. and Swarts, D. C. (2018). Heterologous Expression and Purification of the CRISPR-Cas12a/Cpf1 Protein. Bio-protocol 8(9): e2842. DOI: 10.21769/BioProtoc.2842.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 380
Illegal BglII site found at 1922
Illegal BglII site found at 2231 - 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 196
Illegal PstI site found at 1258
Illegal PstI site found at 3667
Illegal PstI site found at 3865
Illegal NgoMIV site found at 3466 - 1000COMPATIBLE WITH RFC[1000]