Part:BBa_K2934002
pKatA promoter for Bacillus subtilis
pKatA is the B. subtilis catalase promoter and is regulated by the presence of hydrogen peroxide in the medium. At low concentrations of hydrogen peroxide, the PerR protein represses the production of catalase by binding the KatA promoter. When hydrogen peroxide is present, the PerR unbinds
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
The KatA promoter regulates the expression of catalase in Bacillus subtilis. Its repressor, PerR, is a homodimer that has two metal ion binding sites – a structural Zn2+ and a regulatory Fe2+ [1]. When binds its cofactor Fe2+, it represses the expression of the downstream genes by binding the DNA [2]. It can only bind DNA in absence of hydrogen peroxide, as the presence of hydrogen peroxide leads to Fe2+-catalyzed oxidation of histidine residues, which interferes with the repressor's ability to bind the promoter, resulting in the expression of the PerR-regulated gene [3]. Thus, the downstream gene can only be transcribed in the presence of hydrogen peroxide.
Biological Assay
To test the promoter, we have created a vector containing pKatA and mCherry as a reporter gene. This vector was constructed by Gibson assembly, with the pBE-S commercial plasmid (TaKaRa) that served as a backbone (figure 2) and eventually transformed into Bacillus subtilis (RIK1285). The bacteria were cultured in several Erlenmeyer flasks with BioAssay (BA) medium (containing 5% LB), until it reached OD600 of 0.4.
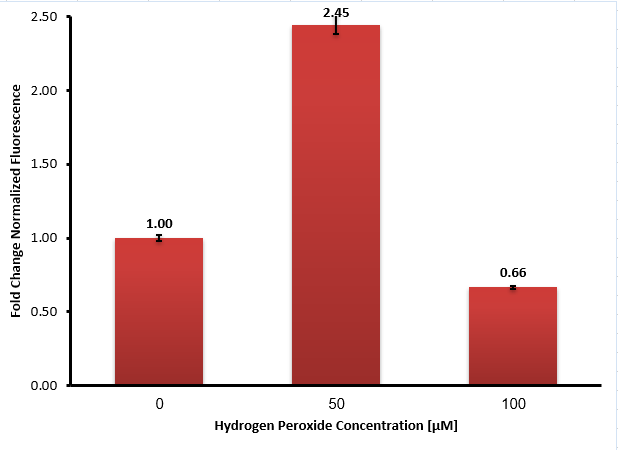
Then, we added different concentrations of hydrogen peroxide to each flask and incubated them for another hour. We measured the fluorescence intensity (at 612 nm) of each sample using plate-reader, and the results are shown in Figure 3:
Our experiment results show that in the absence of hydrogen peroxide high fluorescence intensity was measured, which means the promoter is leaky, although there is a significant increase in the presence of 50 μM hydrogen peroxide. We assume that the higher concentration of hydrogen peroxide (100 µM) has probably disrupted the fluorescence by either interfering with the mCherry molecules or bacteria growth, as seen in the graph as a decrease in the fluorescence intensity.
Methods
For further reading of our full protocols (cloning and the promoter biological assay) look at our wiki protocols page.
.
References:
[1] Mongkolsuk S, Helmann JD. 2002. Regulation of inducible peroxide stress responses. Mol Microbiol 45:9–15.
[2] Lee JW, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367.
[3] Herbig AF, Helmann JD. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol 41:849–859.
| None |