Part:BBa_K2812003
Coding sequence for trunctated Lysostaphin regulated by T7-promoter
This biobrick contains the coding domain for truncated lysostaphin, based on the coding sequence derived from BBa_K748002. In this biobrick, lysostaphin production is regulated by the T7 promoter BBa_K525998. Expression can be induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG). TU-Eindhoven 2018 used this part to produce lysostaphin from Escherichia coli to kill Staphylococcus aureus biofilms for the treatment of wound infections. For more information about our project, please visit our [http://2018.igem.org/Team:TU-Eindhoven wiki].
Usage & Biology
Lysostaphin
Lysostaphin is an antimicrobial agent produced by Staphylococcus simulans. It targets the cell wall peptidoglycan found in certain Staphylococci by cleaving its cross-linking pentaglycine bridges. Among others, it is effective for degrading Staphylocuccus aureus biofilms.1 The encoding part of the lysostaphin has been derived from BBa_K748002, which was made by iGEM Harbin 2012 and was also used by iGEM Stockholm 2016. iGEM Eindhoven 2018 codon optimized this lysostaphin construct. Lysostaphin belongs to the major class of antimicrobial proteins and peptides known as bacteriocins. Bacteriocins are proteins or peptides produced by bacteria, displaying a bactericidal activity against other subpopulations of bacteria.2 The cell wall degradation capability of lysostaphin derives from its endopeptidase activity on pentaglycine cross-bridges in the peptidoglycan layer. Specific cleavage between the third and fourth glycine residue leads to the destruction of the peptidoglycan layer and subsequent lysis of the bacteria.
Experimental Characterisation by TU Eindhoven (2018)
Cloning
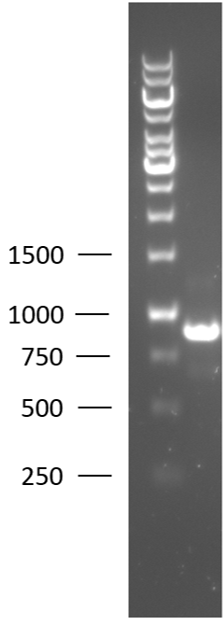
TU-Eindhoven 2018 has characterized the biobrick BBa_K2812003 at the DNA level. First, the T7-lysostaphin construct was synthesized by IDT and subsequently double digested and assembled into the digested linearized pSB1C3 backbone via ligation. The ligated construct was successfully transformed into E. coli NovaBlue, followed by a colony PCR using the VF2 and VR primers to investigate if the correct length has been inserted in the vector. The mixture was ran on a 1% agarose gel as can be seen in figure 1. The observed length of the brightest band corresponds with the expected length of the insert, confirming that the desired construct has been succesfully ligated in pSB1C3 and subsequently transformed in E. coli NovaBlue. Next, the colonies with the correct insert were cultured in LB followed by a plasmid purification by a miniprep. The isolated plasmid DNA was sent for Sanger sequencing and the sequence could be confirmed.
Sources
1) Tossavainen, H., Raulinaitis, V., Kauppinen, L., Pentikäinen, U., Maaheimo, H., & Permi, P. (2018). Structural and Functional Insights Into LysostaphinâSubstrate Interaction. Front Mol Biosci.
2) Bastos, M. d., Coutinho, B. G., & Coelho, M. L. (2010). Lysostaphin: A Staphylococcal Bacteriolysin with Potential Clinical Applications. Pharmaceuticals (Basel), 1139â1161.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 164
- 1000COMPATIBLE WITH RFC[1000]
//cds/enzyme
//cds/enzyme/lysis
//cds/membrane
//cds/membrane/lysis
//chassis/prokaryote/ecoli
//collections/biofilm
//plasmid/expression
//plasmid/expression/t7
//plasmidbackbone/expression/inducible
//plasmidbackbone/synthesis
//proteindomain
//proteindomain/cleavage
//proteindomain/degradation
| None |