Difference between revisions of "Part:BBa K274002"
| Line 6: | Line 6: | ||
[http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] has further characterised ''vio'' operon ([[Part:BBa_K274002]]) and ''vio'' operon ABCE ([[Part:BBa_K274004]]). We have analysed the products of both parts by thinlayer liquid chromatography and mass spectrometry (for a detailed description of the procedures see the 2010 iGEM team Slovenia [http://2010.igem.org/Team:Slovenia/METHODS_and_PARTS/protocols/bm#TLC wiki]). For results of our experiments see the page [https://parts.igem.org/Part:BBa_K274002:Experience experience]. | [http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] has further characterised ''vio'' operon ([[Part:BBa_K274002]]) and ''vio'' operon ABCE ([[Part:BBa_K274004]]). We have analysed the products of both parts by thinlayer liquid chromatography and mass spectrometry (for a detailed description of the procedures see the 2010 iGEM team Slovenia [http://2010.igem.org/Team:Slovenia/METHODS_and_PARTS/protocols/bm#TLC wiki]). For results of our experiments see the page [https://parts.igem.org/Part:BBa_K274002:Experience experience]. | ||
| − | [[Image:SloViolacein-pot.jpg| | + | [[Image:SloViolacein-pot.jpg|center|thumb|300px| '''Figure: The violacein biosynthetic pathway.''' Genes for violacein biosynthesis are arranged in an operon consisting of ''vio''A, ''vio''B, ''vio''C, ''vio''D and ''vio''E. VioA generates an IPA imine from L-tryptophan and VioB converts the IPA imine into a dimer. VioE then acts by transforming the dimer into protodexyviolaceinic acid (PVA), which can be spontaneously converted into a green pigment called deoxychromoviridans. VioD and VioC hydroxylate PVA to form violacein. |
.]] | .]] | ||
Latest revision as of 22:03, 27 October 2010
Vio operon
Produces a purple pigment (violacein) from L-tryptophane. The operon contains five genes (VioA-E) each with their own ribosome binding sites.
[http://2010.igem.org/Team:Slovenia 2010 iGEM team Slovenia] has further characterised vio operon (Part:BBa_K274002) and vio operon ABCE (Part:BBa_K274004). We have analysed the products of both parts by thinlayer liquid chromatography and mass spectrometry (for a detailed description of the procedures see the 2010 iGEM team Slovenia [http://2010.igem.org/Team:Slovenia/METHODS_and_PARTS/protocols/bm#TLC wiki]). For results of our experiments see the page experience.
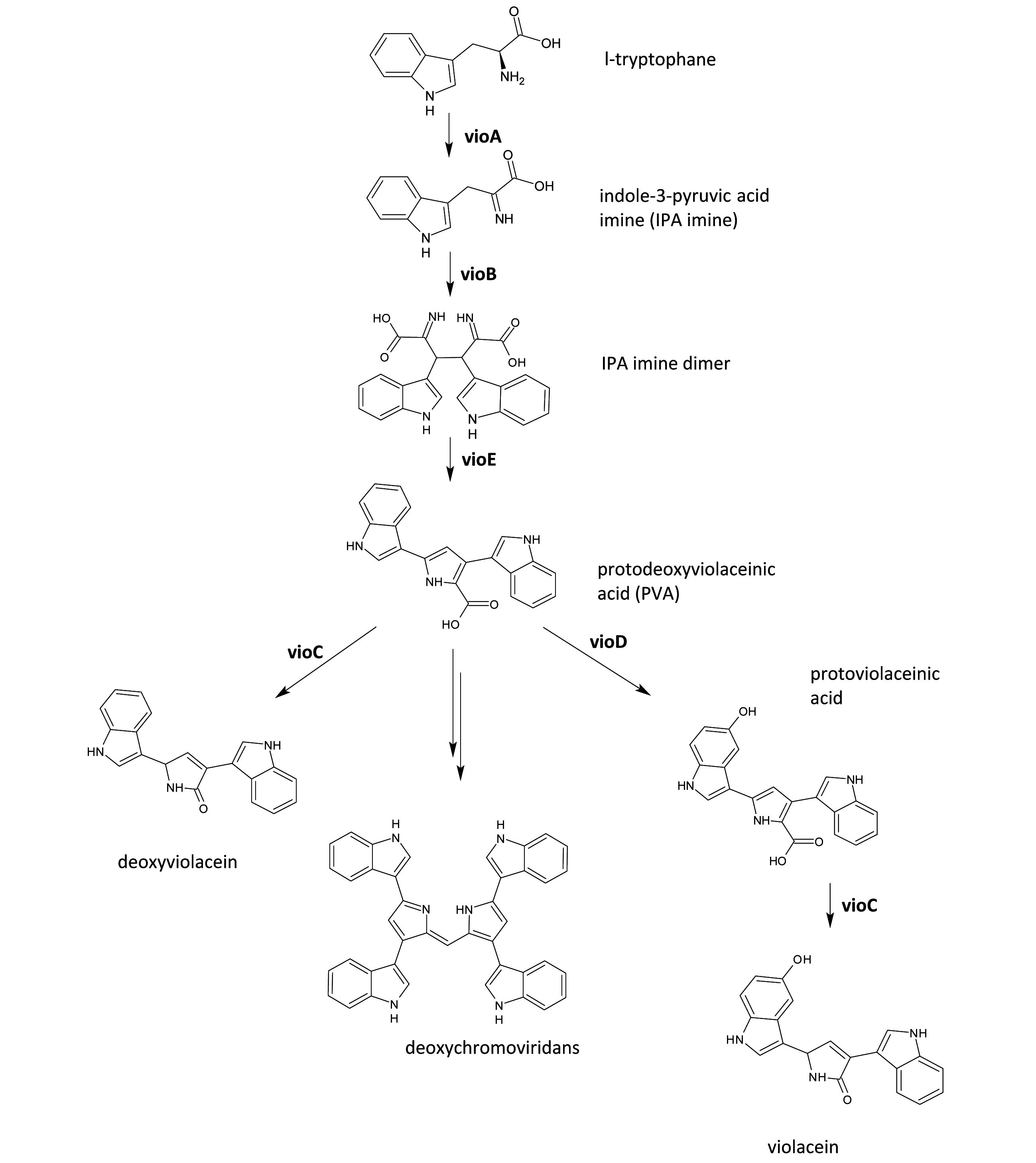
Figure: The violacein biosynthetic pathway. Genes for violacein biosynthesis are arranged in an operon consisting of vioA, vioB, vioC, vioD and vioE. VioA generates an IPA imine from L-tryptophan and VioB converts the IPA imine into a dimer. VioE then acts by transforming the dimer into protodexyviolaceinic acid (PVA), which can be spontaneously converted into a green pigment called deoxychromoviridans. VioD and VioC hydroxylate PVA to form violacein. .
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1029
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 5602
Illegal BamHI site found at 4289 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 3814
Illegal AgeI site found at 4010 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 6552
Illegal SapI.rc site found at 6627
