Difference between revisions of "Part:BBa K2455002"
(→Cell-Penetrating Peptides) |
(→Biobrick Creation) |
||
| Line 34: | Line 34: | ||
The [[part:BBa_K2455002|R9-SYFP2-H6]] biobrick was constructed by amplifying [[Part:BBa_K864100|SYFP2]] using primers with overhangs designed for insertion into the USER cassette. The PCR product was then purified and ligated into the vector, before being restriction digested and sequenced. | The [[part:BBa_K2455002|R9-SYFP2-H6]] biobrick was constructed by amplifying [[Part:BBa_K864100|SYFP2]] using primers with overhangs designed for insertion into the USER cassette. The PCR product was then purified and ligated into the vector, before being restriction digested and sequenced. | ||
| − | == | + | ==Purification and SDS-PAGE== |
| + | The [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K2455002|R9-SYFP2-H6]] proteins were expressed and purified in order to test for internalisation of the proteins in bacteria. All expression were done in BL21 <i>Escherichia coli</i> under control of the T7 promoter. Protein expression was induced with an IPTG concentration of 1.0 mM as optimal expression levels were found at this concentration. | ||
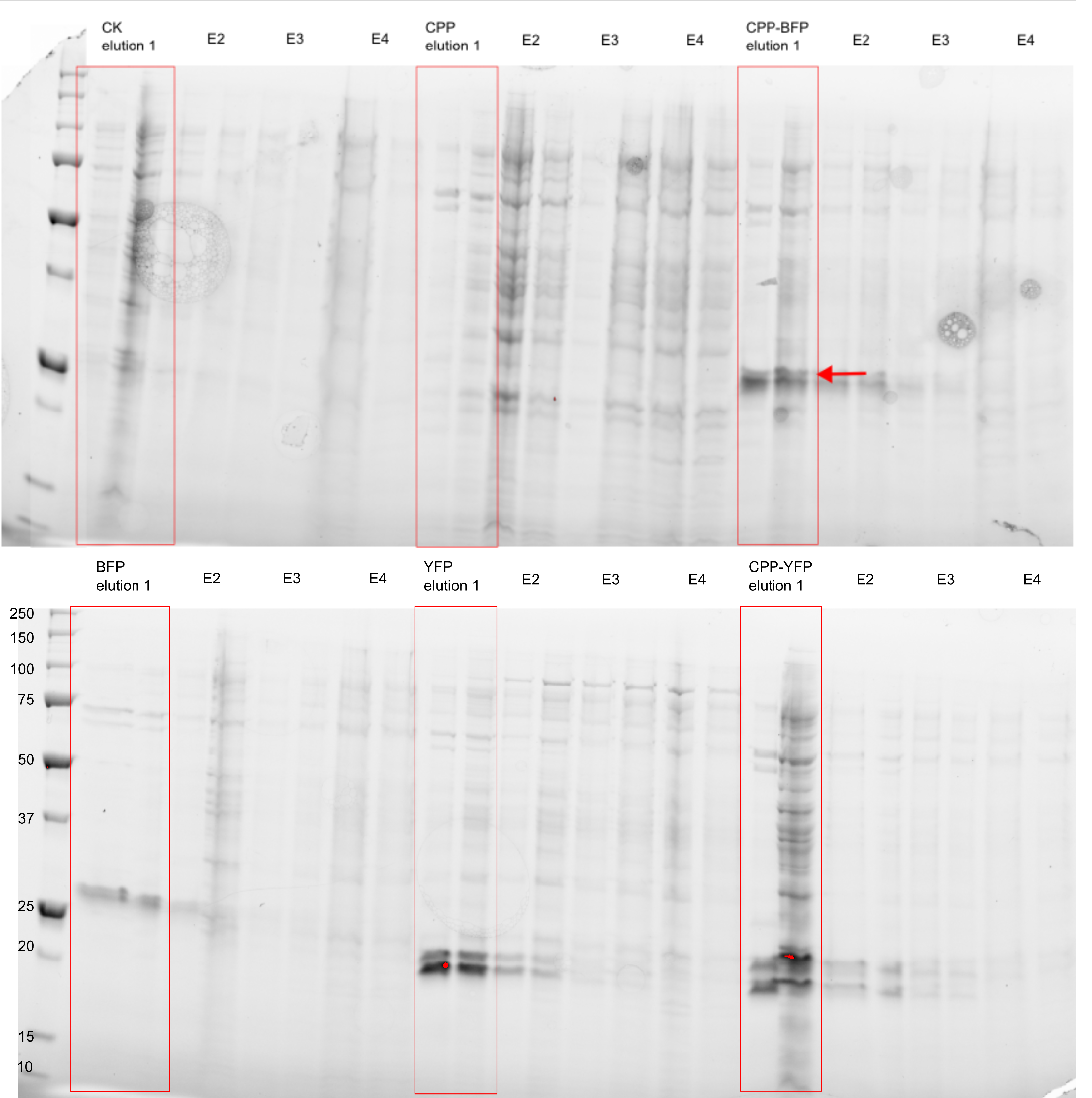
| − | + | [[Image: My_western_image.png|300px|thumb|right|'''Figure 3: Purification of fluorescent proteins.''' Expressed proteins were purified using an Imidazole gradient; E1: 250 mM, E2: 500 mM, E3: 1.0 M, E4: 2.0 M]] | |
| + | |||
| + | ===Purification=== | ||
| + | Cells were harvested, re-suspended in a lysis buffer containing 25 mM Imidazole, 0.5 M NaCl, 20 mM Tris-HCl (pH 7.9), and 1.0 mM PMSF, and lysed by sonication. Cell debris was removed by centrifugation at low speed, and the supernatant was transferred to a fresh falcon tube and mixed with 100 µL [https://www.qiagen.com/us/shop/sample-technologies/protein/expression-purification-detection/ni-nta-agarose/#orderinginformation Ni-NTA Agarose]. The mixture was incubated for one hour on a cold room turning table, where after the beads were spun down and the supernatant removed. A series of elution buffers (0.5 M NaCl, 20 mM Tris-HCl; pH 7.9) with increasing Imidazole concentration was used to elute the bound proteins by consecutively adding 100 µL elution buffer, centrifuging the sample at low speed, and removing 100 µL eluate. | ||
| − | === | + | <b>NOTE:</b> Samples were kept on ice at all times and centrifugation took place at 4 degrees Celsius. |
| + | |||
| + | ===SDS-PAGE=== | ||
| + | The purity of the samples was analysed by subjecting them to an SDS-PAGE using a [http://www.bio-rad.com/en-cn/product/criterion-precast-gels/criterion-tgx-stain-free-precast-gels pre-cast stain free gel]. In addition to [[Part:BBa_K864100|SYFP2]] and [[Part:BBa_K2455002|R9-SYFP2-H6]] two versions of the [[Part:BBa_K592100|mTag BFP]] biobrick were also purified, with an without a CPP-tag. The Imidazole concentration of the purification ranged from 250 mM to 2.0 M, as can be seen on Figure 3 proteins of the expected size are eluted at Imidazole concentrations of 250 mM for all constructs (BFP & YFP 28kDa, CPP-BFP & CPP-YFP 30kDa). | ||
Revision as of 00:37, 2 November 2017
CPP-SYFP2
This is an improved version of the Super Yellow Fluorescent Protein 2 (BBa_K864100) carrying an N-terminal nona-arginin tag, and a C-terminal hexa-histidine tag. The R9-tag is a Cell-Penetrating Peptide giving its associated protein the ability to pass through plasma membranes.
For our study we demonstrated that this improved version of SYFP2 had fluorescence of similar levels to the original protein, and was able to stain both Escherichia coli and Pseudomonas putida cells, presumably through internalisation of the protein.
Previous studies have shown R9 associated proteins to be able to enter a variety of cell types (Chang et al., 2005).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
SYFP2
SYFP2 is a monomeric GFP-based protein giving a bright yellow fluorescence. It has an excitation peak at 515 nm, an emission peak at 527 nm and a relative brightness compared to EYFP2(Q96K) of 12.0 in E. coli.
This version of SYFP2 has an N-terminal R9-tag and a C-terminal H6-tag.
Cell-Penetrating Peptides
Cell-Penetrating Peptides (CPPs) are small peptides, that are able to facilitate transport of a wide variety of cargoes across plasma membranes. CPPs are typically rich in basic residues and originate from viral domains such as the viral HIV tat domain (Eudes & Chugh, 2008). In recent years researchers have tried to make synthetic CPPs, with special interest in peptides constructed solely from arginine residues. The arginine rich sequences have been shown to trigger endocytosis in a wide range of cell types, including onion and potato cells (Chang et al., 2005).
Biobrick Creation
The R9-SYFP2-H6 biobrick was constructed by amplifying SYFP2 using primers with overhangs designed for insertion into the USER cassette. The PCR product was then purified and ligated into the vector, before being restriction digested and sequenced.
Purification and SDS-PAGE
The SYFP2 and R9-SYFP2-H6 proteins were expressed and purified in order to test for internalisation of the proteins in bacteria. All expression were done in BL21 Escherichia coli under control of the T7 promoter. Protein expression was induced with an IPTG concentration of 1.0 mM as optimal expression levels were found at this concentration.
Purification
Cells were harvested, re-suspended in a lysis buffer containing 25 mM Imidazole, 0.5 M NaCl, 20 mM Tris-HCl (pH 7.9), and 1.0 mM PMSF, and lysed by sonication. Cell debris was removed by centrifugation at low speed, and the supernatant was transferred to a fresh falcon tube and mixed with 100 µL Ni-NTA Agarose. The mixture was incubated for one hour on a cold room turning table, where after the beads were spun down and the supernatant removed. A series of elution buffers (0.5 M NaCl, 20 mM Tris-HCl; pH 7.9) with increasing Imidazole concentration was used to elute the bound proteins by consecutively adding 100 µL elution buffer, centrifuging the sample at low speed, and removing 100 µL eluate.
NOTE: Samples were kept on ice at all times and centrifugation took place at 4 degrees Celsius.
SDS-PAGE
The purity of the samples was analysed by subjecting them to an SDS-PAGE using a [http://www.bio-rad.com/en-cn/product/criterion-precast-gels/criterion-tgx-stain-free-precast-gels pre-cast stain free gel]. In addition to SYFP2 and R9-SYFP2-H6 two versions of the mTag BFP biobrick were also purified, with an without a CPP-tag. The Imidazole concentration of the purification ranged from 250 mM to 2.0 M, as can be seen on Figure 3 proteins of the expected size are eluted at Imidazole concentrations of 250 mM for all constructs (BFP & YFP 28kDa, CPP-BFP & CPP-YFP 30kDa).