Part:BBa_K2350004
Contents
NrtA (a part of nitrate channel protein from Synechocystis sp. PCC 6803)
This part codes a part of nitrate channel protein from cyanobacteria Synechocystis sp. PCC 6803 which plays the role of catching nitrate ion from the environment.
Usage and Biology
NrtA is a high-affinity nitrate/nitrite-binding lipoprotein .On Synechocystis sp. PCC 6803, it is tethered to the cell membrane by a lipidated cysteine and a flexible linker rich in glycine and serine.The two domains of it are both composed of five-stranded mixed sheets surrounded by helices. When catch occurs, the nitrate ion is on the middle region of two domains.The resonance state of the nitrate during binding distributes evenly among the three oxygen atoms. However, the second oxygen atom and its interactions with the affinity protein will be absent in the case of nitrite. The second oxygen atom in the bound nitrate molecule also has relatively few interactions. It is the possible answer that nitrate and nitrite have almost the same affinity on NrtA protein.
Analysis
To test whether NrtA protein could capture nitrate and nitrite, we used French Pressure Press to lyse the cell. After dialysis of the cell lysates containing NrtA protein, we used Cayman Nitrate/Nitrite Colorimetric Assay Kit to measure the nitrate and nitrite concentration in the medium.
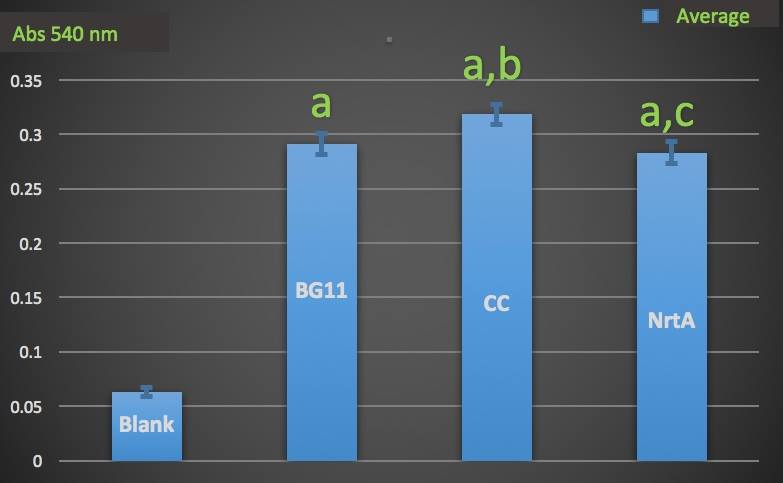
Figure 1.
Nitrate absorbance of different cell lysate.
Blank: nitrate concentration assay kit assay buffer.
CC: competent cell.
BG11: microalgae culture medium buffer.
NrtA: NrtA producing E.coli.
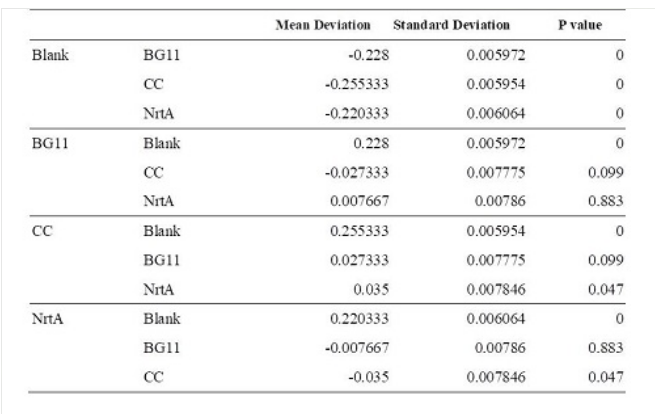
Table 1. Dunnett t test.
Blank: nitrate concentration assay kit assay buffer. CC: competent cell. BG11: microalgae culture medium buffer. NrtA: NrtA producing E.coli.
The nitrate and nitrite concentrations of competent cell and NrtA were significantly different. The results indicated that NrtA protein can capture nitrite and/or nitrate.
Then we wanted to know whether the engineered E.coli could decrease nitrate and nitrite in the medium. We used Cayman Nitrate/Nitrite Colorimetric Assay Kit to measure the nitrate and nitrite concentration of the supernatant of the engineered and native E.coli liquid culture.
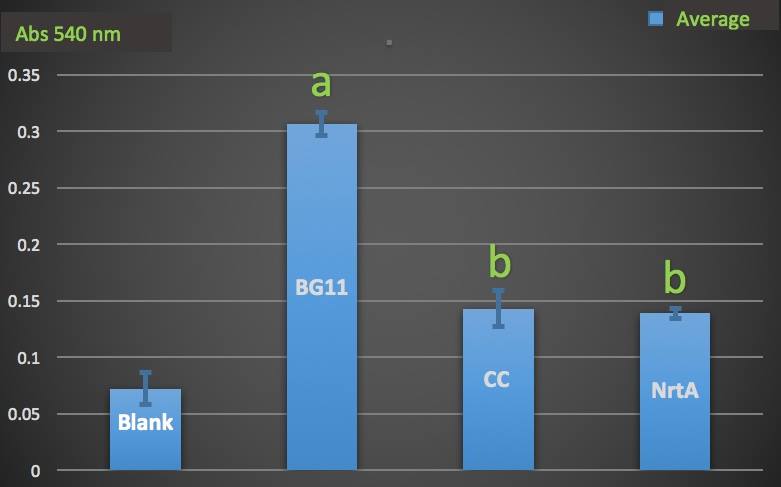
Figure 2. Nitrate absorbance of cell.
Blank: nitrate concentration assay kit assay buffer. CC: competent cell. BG11: microalgae culture medium buffer. NrtA: NrtA producing E.coli.
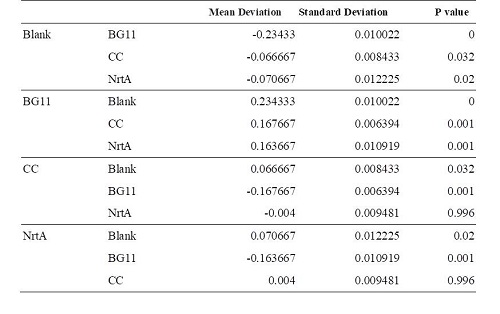
Table 2. Dunnett t test.
Blank: nitrate concentration assay kit assay buffer. CC: competent cell. BG11: microalgae culture medium buffer. NrtA: NrtA producing E.coli.
The nitrate and nitrite concentrations of NrtA and competent cell had slight but not significant difference. The result implied that NrtA protein could capture nitrate and nitrite while the engineered E.coli with NrtA gene could not. The engineered E.coli with NrtA gene could express NrtA protein, and the NrtA protein might be inside the cell, so nitrate and nitrite concentration outside the cell did not change.
References
1. Atomic structure of a nitrate-binding protein crucial for photosynthetic productivity Nicole M. Koropatkin, Himadri B. Pakrasi, and Thomas J. Smith Koropatkin et al.PNAS June 27, 2006 vol. 103 no. 26 9821
2. Substrate-binding Lipoprotein of the Cyanobacterium Synechococcus sp. Strain PCC 7942 Involved in the Transport of Nitrate and Nitrite* Shin-ichi Maeda and Tatsuo Omata. The Journal of Biological Chemistry Vol. 272, No. 5, Issue of January 31, pp. 3036 –3041, 1997
3. Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport Tatsuo Omata, Masayuki Ohmorit, Nobuyuki Arai, and Teruo Ogawa. Proc. Nati. Acad. Sci. USA Vol. 86, pp. 6612-6616, September 1989
This part produces a part of nitrate channel protein from Synechocystis sp. PCC 6803
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1179
Illegal BamHI site found at 549 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |