Part:BBa_K2315006
RpaR-pRpa-GFP
Group: Shanghaitech iGEM 2017
The HSL receiver RpaR from Rhodopseudomonas palustris (strain ATCC BAA-98 / CGA009) activates expression of GFP protein in response to Coumaroyl-HSL.
This Receiver can be easily replaced by other AHL receivers in our collection. A full collection could be found in: http://2017.igem.org/Team:Shanghaitech/Library
Usage and Biology
When AHL is added with concentration higher than a critical value, the constitutively expressed RpaR will bind to the AHL molecule Coumaroyl-HSL, dimerize and bind to the pRpa regulatory sequence to activate GFP expression.
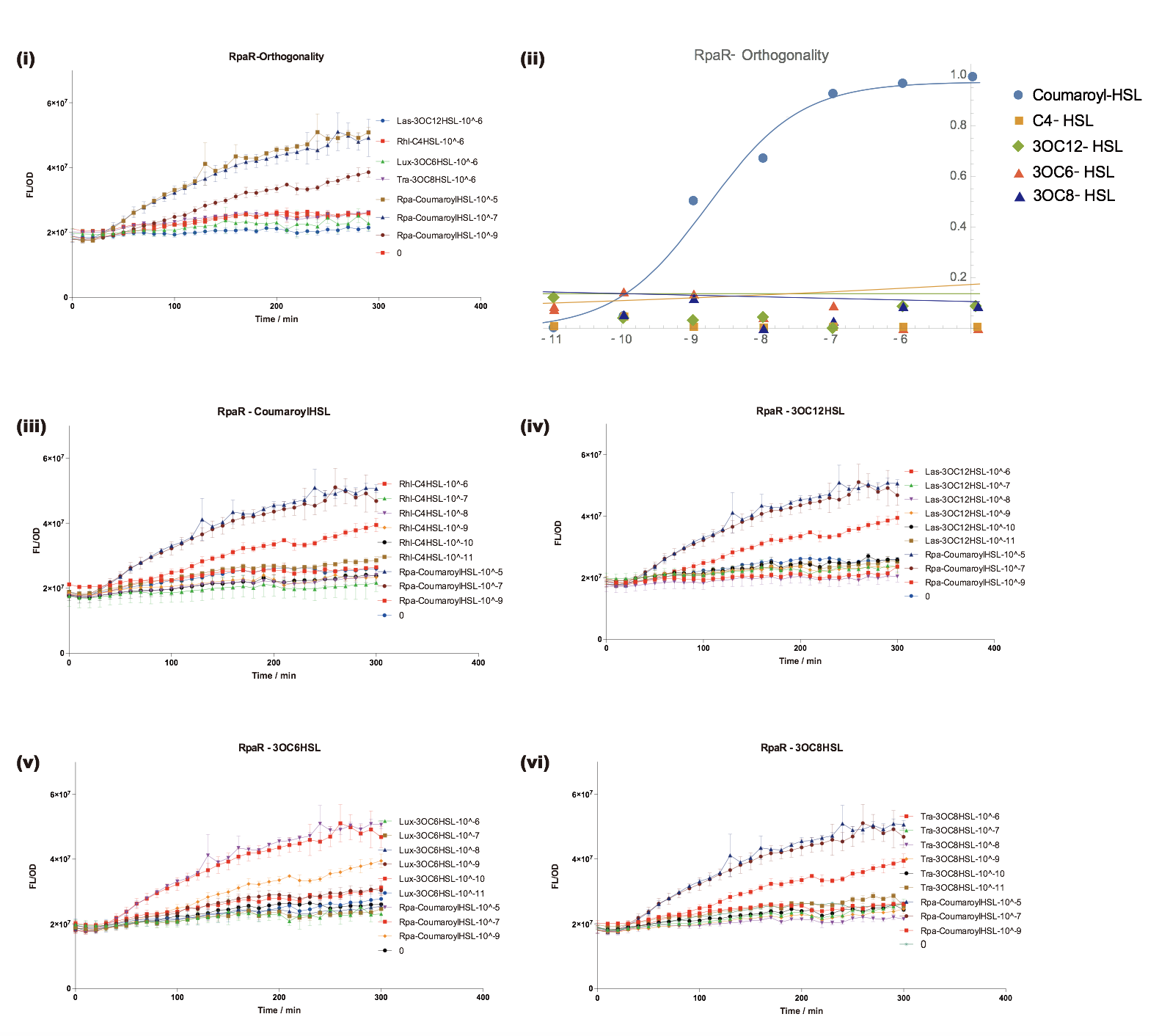
Fluorescent Response to cognate Coumaroyl-HSL
To test this part, we used standard Coumaroyl-HSL (HSL produced by RpaI in P.aeruginosa) to determine the response curve.
Orthogonality test against non-cognate inducers
We have characterized crosstalk response of RpaR to several non-cognate AHLs:
It has shown that RpaR is sensitive to it's cognate HSL and has obvious crosstalk with 3OC8-HSL in Tra System in relatively high concentration.
RpaR-RHOPA-pLas-GFP
RpaR-RHOPA-pLas-GFP
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 493
Illegal XhoI site found at 128 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 136
Illegal NgoMIV site found at 200
Illegal NgoMIV site found at 485
Illegal NgoMIV site found at 573 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1704
| None |