Part:BBa_K2285020
J23119+Target3
This part is just the sequence of our improved Target of Star3 system. Downstream reporter genes are not included so any CDS to be controled can be added just after the Target3.
Usage and Biology
The usage of STAR3 is the same as STAR1.
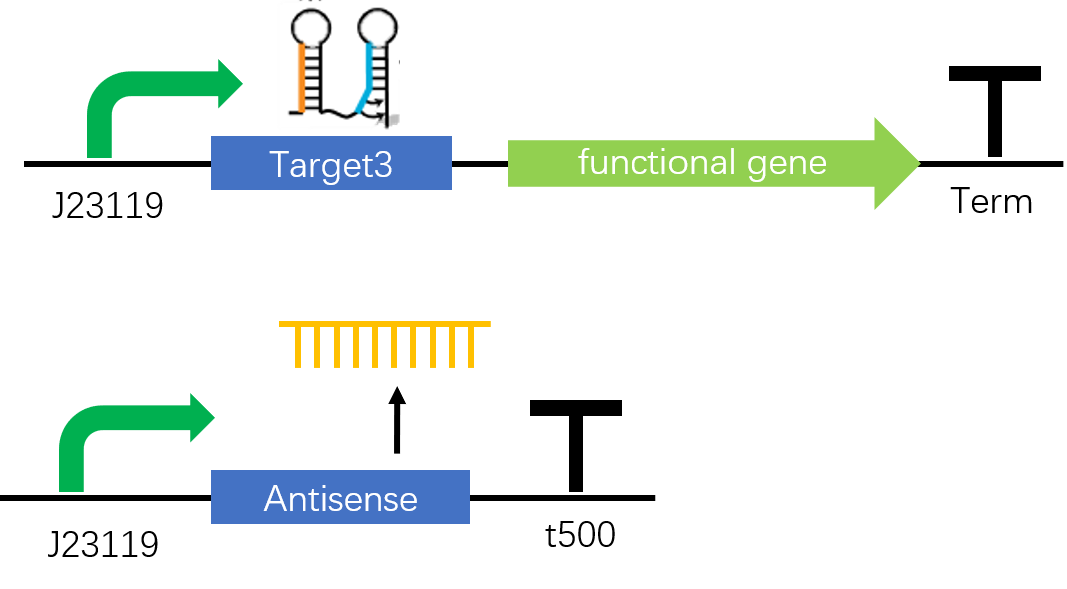
Figure 1. The STAR construct. In the absence of antisense, sense target RNA will form a stem-loop structure, functioning as a terminator and stop the following transcription. When antisense RNA, a mRNA fragment which is the antisense of part of the target RNA, is transcribed, the terminator structure will be disrupted and switch on the inhibited transcription.
A large variety of chromoproteins can be added at the position functional gene. Here we used amilGFP, amilCP, eforRed, cjBlue four chromoproteins in our detection system but actually there are many others you can choose. To response to change of outer environment, what we need to do is just choosing the right promoter on the upstream of Antisense 1 or 3.
Charaterization
To characterize the STAR3 (also STAR1)system, we constructed the circuit below in one plasmid.
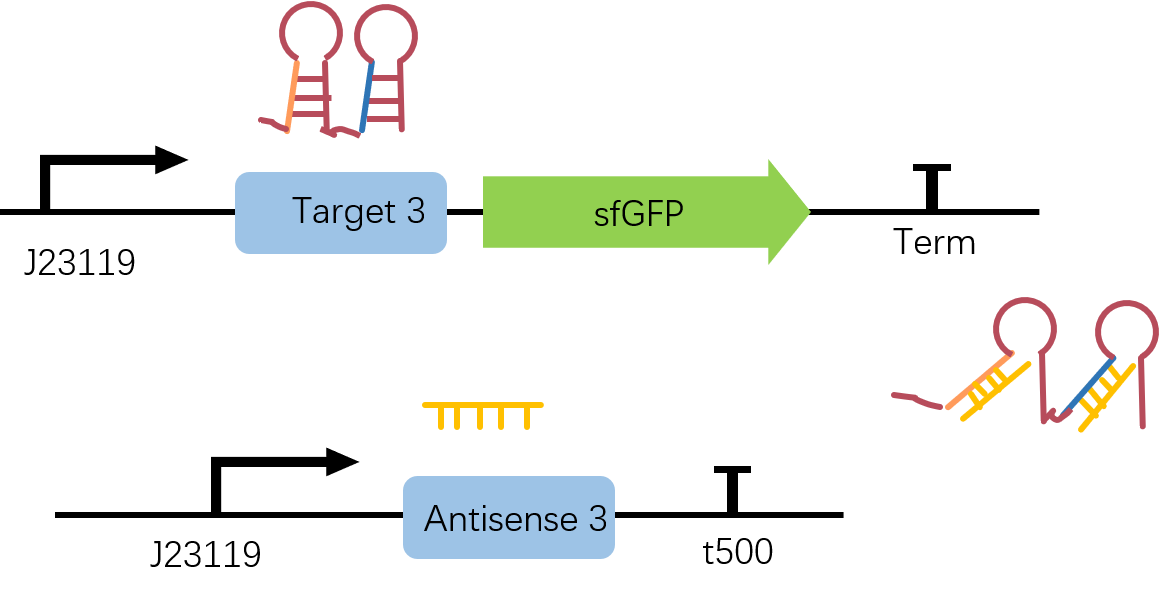
Figure2. Target3-sfGFP & Antisense3 construct. Both Target3 and Antisense3 are on the downstream of constitutive promoter J23119 and two circuits are built in plasmid pET-Duet1.
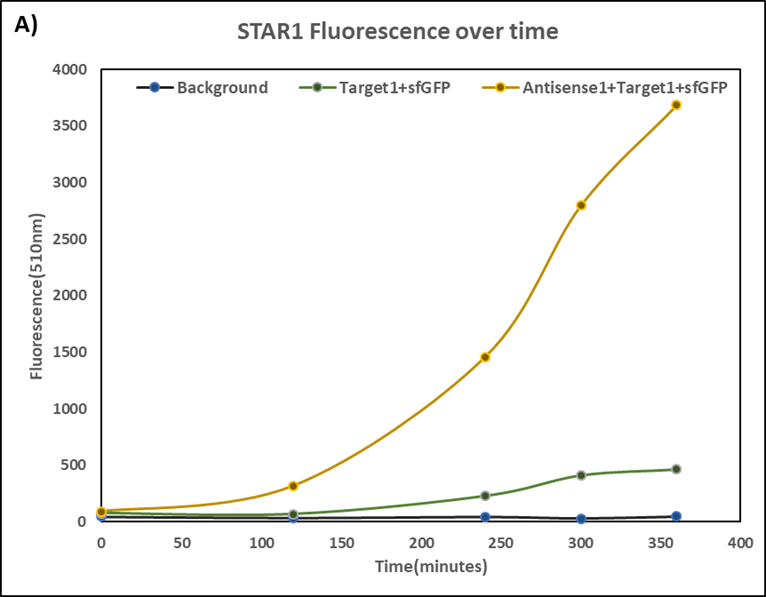
Figure 3. Characterization of STAR1 system.
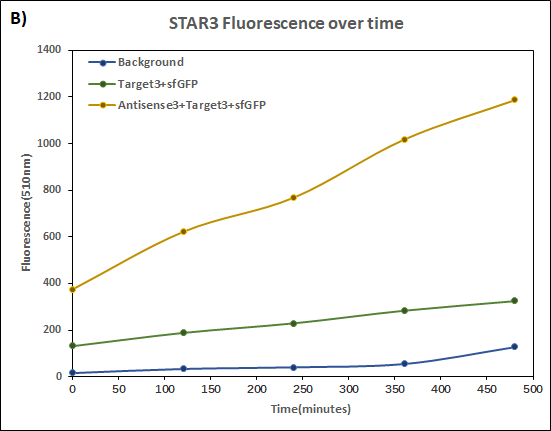
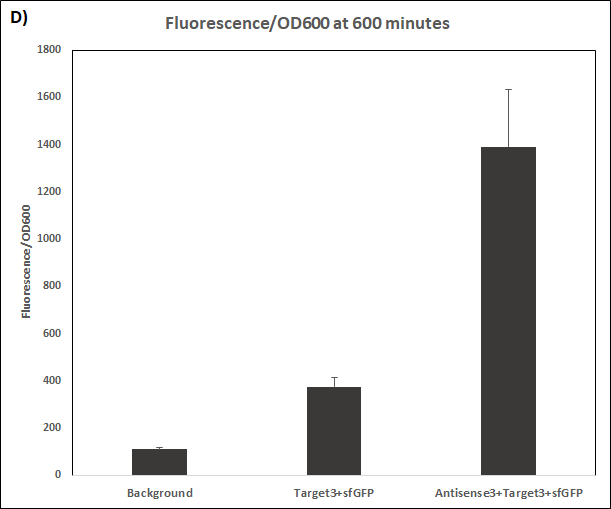
Figure 4. Characterization of STAR3 system.
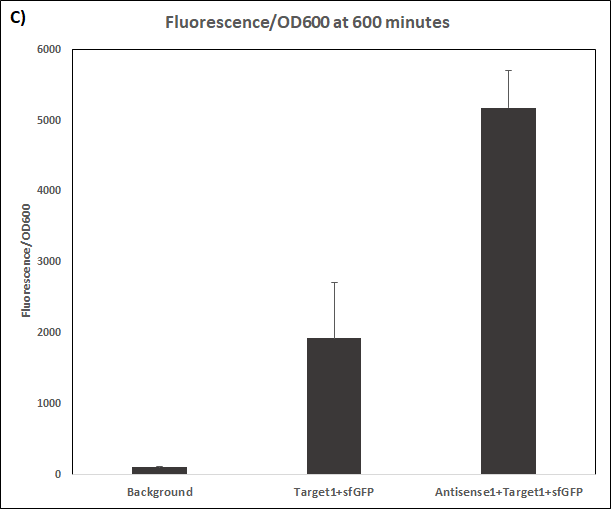
Experiments were performed in E. coli DH5α strain cultured at 37°C. Normalised endpoint fluorescence in the absence or presence of Antisense molecules. We used the dual-expression plasmid pET-Duet1 and pCDF-Duet1 for characterisation experiments involving STARs. For the absence of Antisense, the plasmid did not include Antisense circuit. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH5α cells with only plasmid backbone transformed. Error bars represent standard deviation from 3 technical repeats.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |