Difference between revisions of "Part:BBa K2017004"
| Line 18: | Line 18: | ||
Upstream the luciferase, it is put a linker. In the gRNA testing system, when the device is transcribed and translated, the gene target is attached to the luciferase. When the protein folds, the gene target could interact with the luciferase and affect its tertiary structure. To upstream linker avoids this by creating a distance between the gene target and the luciferase. The linker, theoretically, does not interact with the luciferase. | Upstream the luciferase, it is put a linker. In the gRNA testing system, when the device is transcribed and translated, the gene target is attached to the luciferase. When the protein folds, the gene target could interact with the luciferase and affect its tertiary structure. To upstream linker avoids this by creating a distance between the gene target and the luciferase. The linker, theoretically, does not interact with the luciferase. | ||
| − | This part is one of the standard parts that can be used to create our gRNA testing system device, together with a promoter, terminator and a gene target. | + | This part is one of the standard parts that can be used to create our gRNA testing system device, together with a promoter, terminator and a gene target of your choice. |
<!-- Characterization --> | <!-- Characterization --> | ||
| − | |||
===Characterization=== | ===Characterization=== | ||
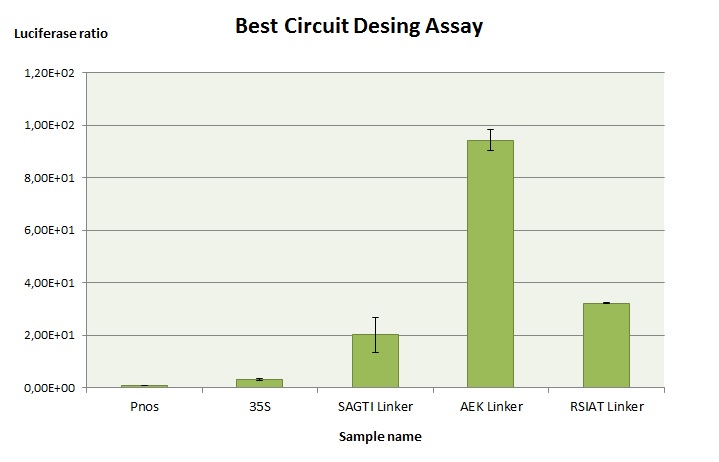
| + | To optimize the gRNA testing system, we tried with three different linkers ([https://parts.igem.org/Part:BBa_K2017003 BBa_K201703], [https://parts.igem.org/Part:BBa_K2017004 BBa_K201704] and [https://parts.igem.org/Part:BBa_K2017005 BBa_K201705]. Our objective was to simulate how well the luciferase would work in the case that the Cas9 would cut the gene target. As the most common editing of the Cas9 according to bibliography was one deletion, for this experiment we designed the target as if it has been cut by the Cas9. This way, the luciferase should be expressed. | ||
| + | |||
| + | The experiment consists on agroinfiltrating the gRNA testing system with a non-useful gRNA (it does not cut the target because we designed it as if it has been cut). Afterwards, a luciferase essay is performed according to our <a href="http://2016.igem.org/Team:Valencia_UPV/Notebook/Protocol#Luciferaseassay_id">protocol</a>. | ||
| + | [[Image:BBa_K2017003.jpeg|600px|thumb|center|'''Figure 1:''' Comparison of ratios between the new gRNA Testing versions. All of them present a higher signal than constitutively expressed luciferase under 35s and pNOS regulation.]] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 19:17, 21 October 2016
RSIAT linker + Luciferase
Firefly luciferase protein. It oxidates its substrate luciferin, emitting light during the process. It can act as a reporter protein when used with a promoter. This part is made to be fused downstream to other coding sequence, so it includes the RSIAT linker in order to let the luciferase acquire the correct tertiary structure.
The luciferase does not include ATG (Met) to initiate translation, as it needs to be fused to other coding sequence on the 5'. The translation must begin in the coding sequence fused upstream. The linker includes a random nucleotide in 5', to change the reading frame of the luciferase. That way, it will not be translated unless an indel is produced in the coding region to which the part is fused.
Sequence and features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 3
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 822
Usage and Biology
Luciferase protein is a useful reporter. It emits light when it oxidates luciferin. In our gRNA testing system, we need to have a truncated luciferase. We have inserted an additional nucleotide before the coding sequence, and we have removed the ATG (Met) that initiates translation. This avoids the luciferase to be expressed unless it is put back to its reading frame. This occurs when it is inserted in our <a href="http://2016.igem.org/Team:Valencia_UPV/Hardware/Reagents#Ourdevice_id">gRNA testing system device</a> and the CRISPR/Cas9 cuts the target. Upstream the luciferase, it is put a linker. In the gRNA testing system, when the device is transcribed and translated, the gene target is attached to the luciferase. When the protein folds, the gene target could interact with the luciferase and affect its tertiary structure. To upstream linker avoids this by creating a distance between the gene target and the luciferase. The linker, theoretically, does not interact with the luciferase.
This part is one of the standard parts that can be used to create our gRNA testing system device, together with a promoter, terminator and a gene target of your choice.
Characterization
To optimize the gRNA testing system, we tried with three different linkers (BBa_K201703, BBa_K201704 and BBa_K201705. Our objective was to simulate how well the luciferase would work in the case that the Cas9 would cut the gene target. As the most common editing of the Cas9 according to bibliography was one deletion, for this experiment we designed the target as if it has been cut by the Cas9. This way, the luciferase should be expressed.
The experiment consists on agroinfiltrating the gRNA testing system with a non-useful gRNA (it does not cut the target because we designed it as if it has been cut). Afterwards, a luciferase essay is performed according to our <a href="http://2016.igem.org/Team:Valencia_UPV/Notebook/Protocol#Luciferaseassay_id">protocol</a>.