Part:BBa_K190028
GlpF
[http://2009.igem.org/Team:Groningen/Project/Transport#GlpF GlpF] is an aquaglycerol porin channel that facilitates the transport of As(III). GlpF is an aquaglycerol porin of E. coli which facilitates not only glycerol import, but also arsenic (As) and antimone (Sb) import (Fu, DX, et al.2000, Meng, YL, et al.2004, Porquet, A, et al.2007, Rosen, BR, et al.2009). It has homologues in other organisms; Fps1p has shown to facilitate arsenic import in yeast and AQP9 is the mammalian homologue (Porquet, A, et al.2007, Rosen, BR, et al.2009). The GlpF aquaglycerol porin is a membrane protein with a symmetric arrangement of four independent GlpF channels. One monomer of this tetramer GlpF porin consists of six transmembrane and two half membrane-spanning α-helices that form a right-handed helical bundle around the channel. The channel has a diameter of ~15Å at the periplasmid end, which constricts towards a diameter of ~3.8Å at the beginning of a 28 Å long selective channel that ends at the cytoplasmic end (Fu, DX, et al.2000). The GlpF is a stereospecific channel that is thought to be more selective on molecular size than on chemical structure (Fu, DX, et al.2000, Heller, KB, et al.1980). It does allow transport of a variance of polyhydric alcohols, glycerol being one of them, and arsenic and antimone (Fu, DX, et al.2000, Meng, YL, et al.2004, Porquet, A, et al.2007,Rosen, BR, et al.2009, Heller, KB, et al.1980). Carbon sugars and ions are shown to be unable to be transported by GlpF (Heller, KB, et al.1980). At physiological pH arsenic and antimone are not present in their ionic state but rather as As(OH)3 and Sb(OH)3 (Rosen, BR, et al.2009). These elements show a charge distribution similar to glycerol and a smaller but comparable volume. The structural similarities are thought to be the reason for the possibility of these elements to be transported in the cell by GlpF (Porquet, A, et al.2007). If GlpF behaves as a nonsaturable transporter, a transport rate of 1µmol of glycerol is transported per minute per mgr of cell protein (Heller, KB, et al.1980).
references:
- [http://2009.igem.org/Team:Groningen/Literature#Meng2004 Meng 2004]
- [http://2009.igem.org/Team:Groningen/Literature#Rosen2009 Rosen 2009]
- [http://2009.igem.org/Team:Groningen/Literature#Porquet,A,etal2007 Porquet, A, et al.2007]
- [http://2009.igem.orgTeam:Groningen/Literature#Fu,DX,etal2000 Fu, DX, et al.2000]
- [http://2009.igem.orgTeam:Groningen/Literature#Heller,KB,etal1980 Heller, KB, et al.1980]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 283
- 1000COMPATIBLE WITH RFC[1000]
Results
Metal sensitivity assay
The ability of GlpF (overexpressed under IPTG induction) to transport As(III) was tested by an arsenite uptake [http://2009.igem.org/Team:Groningen/Protocols assay]. Also the full accumulation device (BBa_K190038) was tested using this assay. Data and analysis can be found [http://2009.igem.org/Team:Groningen/Project/Accumulation here].
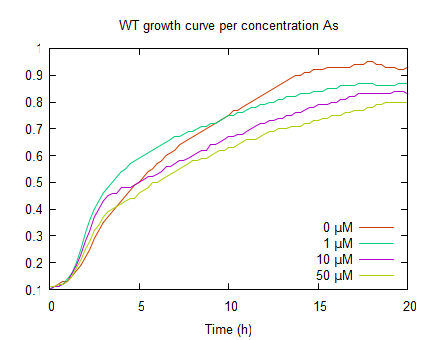
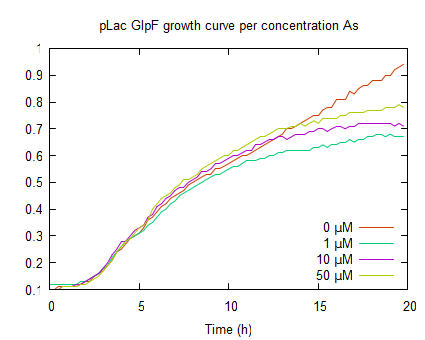
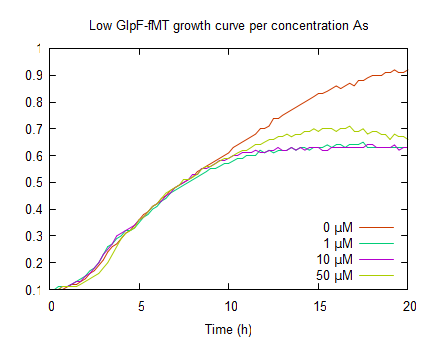
The graphs above represent the result of the metal sensitivity [http://2009.igem.org/Team:Groningen/Protocols#Death_assay assay]. The lines in the graphs represent the average optical density (OD600) of a construct over time. The graph on the left shows that increased As(III) levels inhibits growth and as the As(III) concentration increases the plateau OD600 level decreases.
The second graph represents the OD600 values of a E. coli culture with pLac-GlpF. The growht curves are less steep in the log phase compared to WT, because of the protein expression by IPTG induction. In the absence of As(III) the plateau level equals the WT, but if arsenite is present the plateaus are lower (OD600 <0.8) which implies functional expression of GlpF causing As(III) uptake to toxic levels.
In the graph on the right, shows growth curves of E. coli expressing GlpF and fMT under a low constitutive promoter. Also a similar growth in the log phase, compared to pLac GlpF, can be seen this is due to protein expression. Furthermore, like WT (0μM As(III)) it has its plateau above OD600 0.9. Upon incubation with various arsenite concentrations, the plateaus are lower (OD600 <0.8) compared to WT. This is due to As(III) uptake by GlpF. The reduced growth is also an indicator for arsenite uptake. It is difficult to see if fMT has any effect because this assay can not show where the arsenite is and how fMT interferes with the cells' detoxification.
Arsenic uptake assay
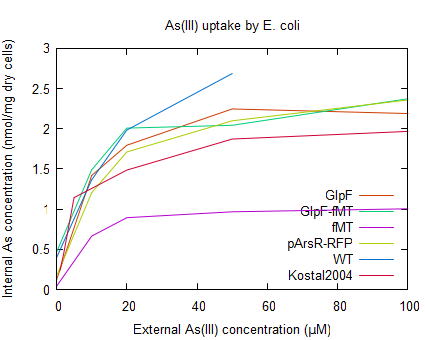
Arsenite uptake by E. coli containing pSB1A2-R0010-GlpF (BBa_K190032) was tested using a arsenic uptake assay (according to Kostal 2004) and arsenic concentration determination by ICP-MS (see [http://2009.igem.org/Team:Groningen/Protocols#Metal_uptake_assay_for_E._coliKostal_2004 protocols]). But because of a lack of reproducibility and an unexpected high uptake yield (as can be seen in figure 4), the improved uptake of arsenite by GlpF could not be determined using this assay. For further reading on discussion of this data see the [http://2009.igem.org/Team:Groningen/Accumulation wiki]
- Figure 4: Uptake of As(III) by E. coli WT, and the strains containing the different parts of the accumulation device. As a control the arsenic uptake of E. coli with ArsR overexpression (as described by Kostal 2004) is also shown.
references:
- Kostal 2004
- Jan Kostal, Rosanna Yang, Cindy H. Wu, et al (August 2004). "[http://dx.doi.org/10.1128/AEM.70.8.4582-4587.2004 Enhanced Arsenic Accumulation in Engineered Bacterial Cells Expressing ArsR]". Applied and Environmental Microbiology 70(8): 4582–4587
Functional parameters
Based on our analysis of data from [http://2009.igem.org/Team:Groningen/Literature#Meng2004 Meng2004]:
| Parameter | Value |
|---|---|
| vmax | > 3.19 µmol/(s·L) |
| K | ± 8.70·vmax |
Optimization
Washington iGEM 2021 referenced the sequence of glpF described in this page for use in our project. We optimized the codons for expression in E.coli (our host organism) using IDT codon optimization tool. According to an analysis by Benchling Codon optimization tool, the optimized glpF had no rare codons (from 3), reduced uridine content from 31% to 29%, while maintaining a GC content at 51%.
//cds/membrane/transporter
| n/a | GlpF |