Part:BBa_K1614000
T7 promoter for expression of functional RNA
Minimal promoter derived from T7 phage promoter.
Usage and Biology
This T7 promoter derivate is especially useful for the use in in vitro applications. The G which is the startpoint for transcription is included in the promoter sequence.
UBrawijaya iGEM 2021
|
UBrawijaya iGEM 2021 project CHOP systemis an outer membrane vesicle based protein overexpression system that includes three goals towards producing a blueprint for an ideal protein production system. This project can be customized because the system can be adapted with various proteins of interest, the overexpression system that can increase protein yield, and the clean harvest with simplified purification steps that is equipped with a protease enzyme expression mechanism in the concentrated system - from vesicles and cell pellets. |
This part is used to tune the expression level of constitutively expressed parts. We decided to characterize two different primary protein-generators to our system; Constitutive promoter family member-RBS and T7 promoter.
Usage & Biology
T7 bacteriophage polymerase as the major gene product of T7 phage has high and specific processivity with a single subunit structure and has the capability of transcribing complete transcriptional cycle without additional proteins. T7 bacteriophage polymerase exhibits an efficient elongation. It elongates about five times faster than E. coli RNA polymerase and. These properties add advantage to this enzyme and are hence widely employed to express heterologous genes, under the control of the T7 promoter [1]. The T7 promoter has high specificity for initiating transcription, DNA binding occurs from -17 to -5 nucleotide positions and the double-stranded DNA is melted from -4 to +3 positions to prime RNA synthesis of the GTP nucleotides at the +1 position [2]. T7RNAP can be inhibited by changing physiological conditions, such as pH, salt concentration, and temperature.
References
[1] Borkotoky, Subhomoi, and Ayaluru Murali, "The highly efficient T7 RNA polymerase: a wonder macromolecule in biological realm." International journal of biological macromolecules 118, Pp:49-56. Oct 2018.
[2] Conrad, Thomas, Izabela Plumbom, Maria Alcobendas, Ramon Vidal, and Sascha Sauer, "Maximizing transcription of nucleic acids with efficient T7 promoters," Communications biology 3, no. 1, Pp: 1-8, Aug 2020.
Characterize the T7 promoter – CCU-Taiwan
In our pHMT-LbCas12a construct, we included the T7 promoter (Part:BBa_K1614000) to induce the LbCas12a expression. To characterize the ability of T7 promoter to induce functional RNA, we examined whether the T7 promoter induced LbCas12a RNA can be translated into proteins by SDS-PAGE and Coomassie blue staining. We first transformed pHMT-LbCas12a into E.coli BL21(DE3), which harbors a Lac Operon regulated T7 polymerase. We then expanded the transformed BL21(DE3) cells by TB medium at 37℃. When O.D.600 approach 0.6 ~ 0.8, we induced T7 polymerase expression by adding 0.2 mM IPTG to TB medium and shift BL21(DE3) to 16℃ for 16 hr.
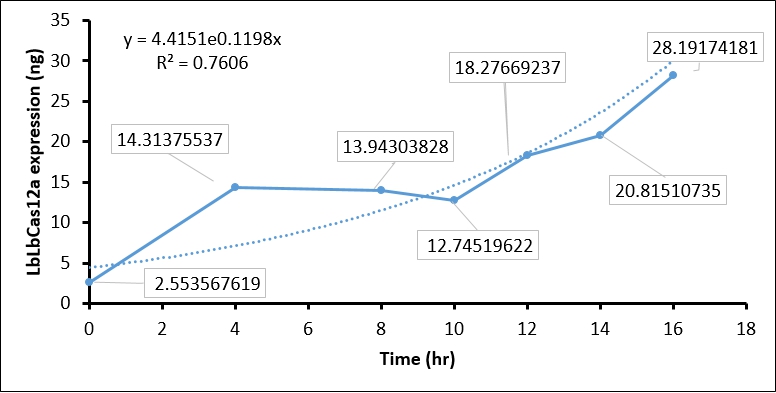
The IPTG treatment increased the T7 polymerase expression in BL21(DE3) as time goes on, which in turn activate T7 promoter. The activated T7 promoter should promote LbCas12 transcription and translation. To examine this, we collected IPTG-induced BL21(DE3) every two hour, and performed SDS-PAGE assay and Coomassie Blue staining to detect LbCas12a protein expression. In brief, 1 ml of BL21(DE3) is harvested every two hour and centrifuged to cell pellet. The cell pellet was then sonicated, and soluble fraction in supernatant was collected by centrifuging at 13200 rpm 30 min. Total 1 ml supernatant, which equal to protein expression in 1ml BL21(DE3), was used in SDS-PAGE and Coomassie Blue staining. To calculate the protein expression at different time points, we quantity the image intensity of LbCas12a protein and BSA standard by Image J.
- Coomassie blue staining shows the LbCas12a protein expression was absent at 0 hr and increased as time goes on. This result indicated that T7 promoter can be activated by IPTG-induced T7 polymerase (Figure 1).
- We then used BSA standard to quantify the LbCas12a protein expression. The simple linear regression of BSA protein expression standard is shown in Figure 2, while the original image intensity detected by Image J is shown in table 1.
- Finally, we quantify the LbCas12 expression by conversing the intensity to concentration through BSA standard regression (Figure 3).
| BSA (ng) | Background intensity | BSA intensity | BSA - Background intensity |
| 350 | 2407241 | 1258862 | 1148379 |
| 210 | 1589271 | 962468 | 626803 |
| 70 | 1544912 | 904623 | 180999 |
| 35 | 1042670 | 861671 | 129877 |
| Time (hours) | Background intensity | LbCas12a intensity | LbCas12a - Background intensity | LbCas12a (ng) | LbCas12a (µg/mL) |
| 0hr +IPTG | 696645 | 691258 | 5387 | 8.937 | 2.554 |
| 4hr +IPTG | 900536 | 760105 | 140431 | 50.098 | 14.314 |
| 8hr +IPTG | 925618 | 789444 | 136174 | 48.801 | 13.943 |
| 10hr +IPTG | 913959 | 791540 | 122419 | 44.608 | 12.745 |
| 12hr +IPTG | 956220 | 770282 | 185938 | 63.968 | 18.277 |
| 14hr +IPTG | 976568 | 761481 | 215087 | 72.853 | 20.815 |
| 16hr +IPTG | 1064337 | 764543 | 299794 | 98.671 | 28.192 |
| 16hr Ctrl | 924162 | 743361 | 180801 | 62.403 | 17.829 |
Conclusion:
The quantification of LbCas12a protein expression by BSA standard shows that the Cas12 protein is increased at different time points, which confirmed that the T7 promoter is functional in our construct.
iGEM Marburg 2021 - Contribution
Cell-free transcription and translation systems are advanced in vitro used as prototyping platforms for metabolic networks and gene circuits. Typically, they are based on crude cell extracts and contain the whole machinery needed for protein biosynthesis. This advantage in cell-free systems offers exciting opportunities to fundamentally transform synthetic biology. It enables new approaches for model-driven design of synthetic gene networks, rapid and portable acquisition of targeted components, as well as on-demand biomanufacturing, and building synthetic cells from scratch. Recently, cell-free technology approaches have gained more and more importance in the synthetic biology field. While the majority of cell-free systems are based on bacterial and eukaryotic cells, there are almost no cell-free systems based on plants. For this reason, our project was focused on developing novel cell-free systems of chloroplasts from various plant species. With our Chloroplast Cell-Free system we provide a unique test-platform, which can be easily applied to study various genetic constructs for plant engineering in a much shorter time frame.
The DNA concentration response of the T7 polymerase in a chloroplast cell-free system
Despite many advantages and simplicity of the system, significant differences in expression using cell-free systems can be caused by the addition of different amounts of template DNA[1]. For this reason, we decided to test the behavior of our system to different DNA concentrations.
In the following experiment, NanoLuc luciferase was used as a reporter system, within our cell-free expression measurements. The graph below indirectly shows the expression levels of this NanoLuc luciferase via the emitted luminescence (see Figure 4). To analyse the influence of DNA concentration to the target protein expression level, extracts from two different plant species (N. tabacum and S. oleracea) have been used and DNA concentrations in the spectrum from 0.5 nM to 15 nM have been added to the final cell-free reaction mix.
For both N. tabacum and S. oleracea, an optimum expression level is reached at DNA concentration of 5 nM. With a further increase of the concentration, no significant differences in expression levels can be recognised.
These results show that it is essential to carefully normalize DNA concentration for the comparison of various parts in the experiments using cell-free systems. Additionally, using saturated DNA concentrations has the advantage of being less prone to variations in expression, caused by the use of different concentrations in DNA.
Moreover it should be “good practice” not to have DNA concentrations as a limiting factor in your measurements, as this could cover other effects/properties one would like to actually characterize in the experiment.

References
[1] Kopniczky, M. B., Canavan, C., McClymont, D. W., Crone, M. A., Suckling, L., Goetzmann, B., Siciliano, V., MacDonald, J. T., Jensen, K., & Freemont, P. S. (2020). Cell-Free Protein Synthesis as a Prototyping Platform for Mammalian Synthetic Biology. In ACS Synthetic Biology (Vol. 9, Issue 1, pp. 144–156). American Chemical Society (ACS). https://doi.org/10.1021/acssynbio.9b00437
//chassis/prokaryote/ecoli
//direction/forward
//plasmid/expression/t7
//promoter
//regulation/constitutive
//rnap/bacteriophage/T7
| None |