Part:BBa_K1467101
GoldenGate compatible 35s Promoter
35S is a plant specific promoter obtained from the Cauliflower Mosaic Virus. The part is intended for use as a constitutive promoter for gene expression experiments in plants. This part is compatible with GoldenGate MoClo Assembly Standard as it is free from internal BsaI and BpiI recognition sequences.
iGEM14_NRP-UEA-Norwich have used this part in constructs to constitutively express proteins of interest, often providing a control to test the expression of each of our reporters. This allowed us to compare expression levels in contrast to when using promoters induced by external stimuli (BS3, PDF1.2, PR1), allowing us to select the most effective inducible promoters. Examples of the way in which the 35s promoter was used in The Green Canary project are shown below.
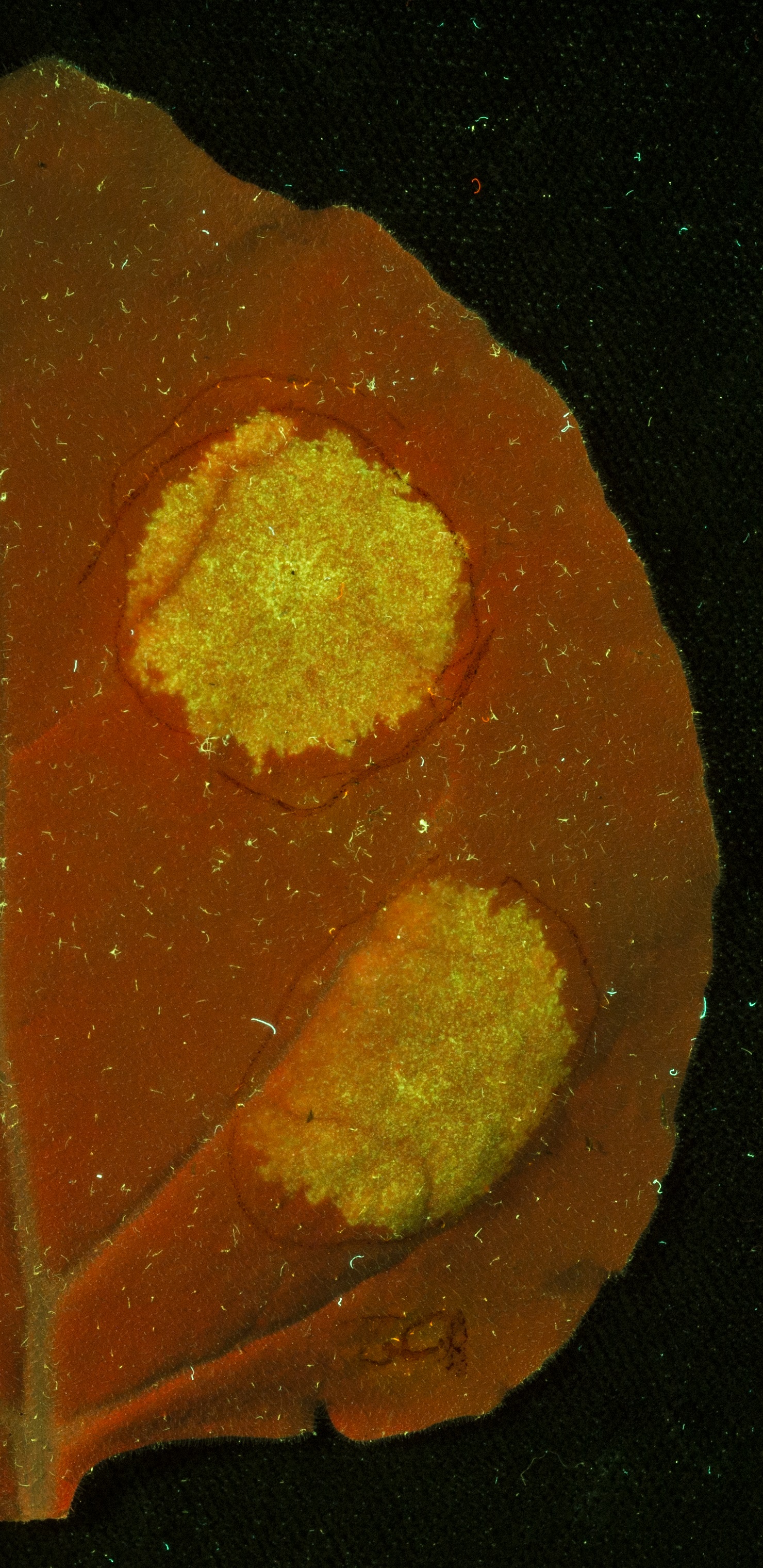
Figure 1: The result of an infiltration of Agrobacterium tumefaciens into Nicotiana benthamiana, showing plant cell apoptosis caused by expression of Bax. This Bax expression also gives evidence for the function of the 35S promoter which is constitutive and therefore causes expression of the protein without external stimuli.
Image (A) provides a control, showing a Nicotiana benthamiana leaf infiltrated (red circles) with Agrobacterium tumefaciens whilst B shows a Nicotiana benthamiana leaf infiltrated (red circles) with Agrobacterium tumefaciens carrying the plasmid encoding: 35s Constututive Promotor (Cauliflower Mosaic Virus) + Bax Coding Sequence (Mus Musculus) + Octopine Synthase Terminator (Agrobacterium tumefaciens).
Figure 2: A Nicotiana benthamiana leaf under UV light following infiltration (red circles) with Agrobacterium tumefaciens carrying the plasmid encoding 35s Constututive Promotor (Cauliflower Mosaic Virus) + GFP Coding Sequence + Octopine Synthase Terminator (Agrobacterium tumefaciens). The result shows GFP expression, driven by the constitutive 35s promoter.
The part is similar to parts BBa_K509000 and BBa_K414002 submitted by previous iGEM teams but has been domesticated to fit the GoldenGate MoClo Assembly Standard. The part can be used by future iGEM teams using the GoldenGate standard, allowing cloning in a one-pot, one-step, digestion-ligation reaction as detailed by the NRP-UEA iGEM 2014 team protocols.
NRP-UEA 2015
At NRP-UEA, we aimed to produce a prebiotic to prevent colon cancer. To do this, we attempted to acylate/butrylate starch in plants. Methods to chemically acylate starch purified from plants already exist, but as they use strong chemicals and require heating they are not environmentally friendly. Using various acyltransferases, we hope to acylate starch in plants. We’ll be using a model plant, Nicotinia benthamiana, for initial tests because we can get results within a few days. We used Golden Gate Cloning and the Plant Standard Syntax described in RFC106 and Patron et al (2015) to make our constructs.
All of our constructs contained the 35s Constitutive Promoter. This was done to drive the constitutive expression of our parts in the plants. We also added a chloroplast transit peptide, acyl transferase, and a 35s terminator to our constructs (BBa_K1618033‐036). In a second set of parts, we added a fluorescent tag, to confirm the function of our chloroplast transit peptide (BBa_K1618029‐032).
We transformed our constructs into Agrobacterium tumefaciens and then infiltrated it into our plants. We used the YFP tagged constructs to confirm the localisation of parts to the chloroplast (Figure 1) and the untagged constructs to analyse the starch content of our leaves.
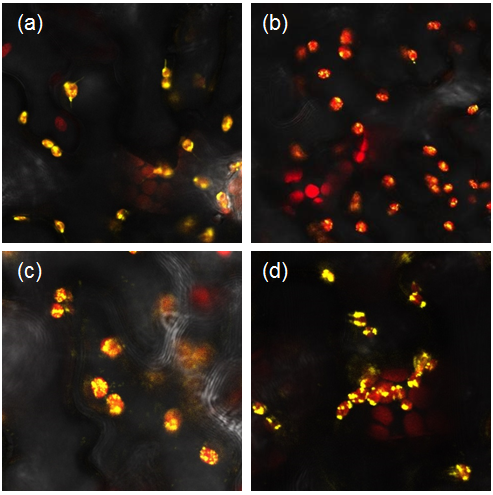
Figure 1: Constructs BBa_K1618029‐032 contain a yellow fluorescent protein, as well as a chloroplast transit peptide. These are confocal microscopy images of the constructs infiltrated into Nicotiana benthamiana, in which the red structures are the chlorophyll within the chloroplast, and the yellow is the fluorescent fusion protein expressed from constructs (a) BBa_K1618029, (b) BBa_K1618031, (c) BBa_K1618032, and (d) BBa_K1618030.
The above results not only indicate that our chloroplast transit peptide works, but also suggest that the 35s promoter that we used is GoldenGate compatible and functioning.
SZ-SHD 2023
This year our team aims to develop a "phytosensor" that revolutionizes the way we monitor phosphate levels within plants. (https://2023.igem.wiki/sz-shd/description#design) To achieve this, we integrated our patented carbon nanodot-based tracked, transformation, translation, and trans regulation(TTTT) technique (https://2023.igem.wiki/sz-shd/plant) and a multi-level gene circuit designed by our team.
Therefore, the CAMV 35S plant promoter was used throughout our whole project. There are some key introductions to our application.
CAMV 35S as a promoter for our next-generation Plant report gene eyGFP_UV and used as a positive control The traditional equipment to detect GFP is not only highly expensive but also requires lots of training for operation. Thus, we are excited to introduce a new plant report gene, called eYGFP-uv (a GFP variant, BBa_K4844000), in transient expression we observed bright fluorescence under UV light on tobacco leaves. eyGFPuv was optimized for maximal fluorescence to be observed by naked eyes under UV light instead of using a fluorescence microscope.
Our transient expression vector was built up with a plant-strong promoter CAMV 35S, a 5'UTR on the upstream of eygfp (uv) to enhance the translation efficiency in plants and fused with a 3*flag tag downstream for western blot detection of the protein. (Our vector sequence can be downloaded at the supplementary material page https://2023.igem.wiki/sz-shd/experiments)

Once the tobacco plant has been transformed with carbon nanodots for three days, bright florescent can be visualized with the naked eye under the light source of a 398nm UV flashlight. We also ran a western blot using the protein extraction of tobacco tissue samples and anti-flag-tag antibodies. A clear band can be observed on the membrane (27.9 kDa).

More information about this application ca be found at: https://2023.igem.wiki/sz-shd/engineering#characterization
Recombined CAMV 35S work in our amplifier gene circuit In order to reduce the leaking expression (background expression without low phosphate pressure) of the original low phosphate promoter we used and increase the promoting strength. We decided to design a signal amplification system.
In our gene circuit, a recombined 35s promoter was used for the out put of our report gene.

Our qPCR result indicates that our low noise amplifier part can not only increase the expression strength but also reduce leaky expression.

More about this design can be found at: https://2023.igem.wiki/sz-shd/engineering#construction
Other potential applications of 35S: Based on our carbon nanodot-based tracked, transformation, translation, and trans regulation(TTTT) system, more potential applications can be done. Lear more at: https://2023.igem.wiki/sz-shd/plant
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 392
Illegal BglII site found at 1138
Illegal XhoI site found at 1342 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//promoter
//regulation/constitutive
| None |