Part:BBa_K1460002
RBS + GST (glutathione-S-transferase)-CRS5 (metallothionein) + Ter
This part serves as a functional metallothionein for heavy metal binding and conferred resistance to heavy metals when combined with an appropriate promoter. CRS5 gene is the metallothionein that binds to heavy metals and it is fused to GST for stability.
Characterization, Improvement and Functional Tests of K1460002
Group: TAS_Taipei 2019
Author: Allison Kuo, Anna Chang and Yasmin Lin
Summary: We inserted part BBa_1460002 into our composite part BBa_K2921550. Our composite part added a colored protein on the end of BBa_K1460002 in order to see the fusion protein once it is bound to Nickel ions.
Characterization
We used SDS-PAGE to check for Met-GS-mRFP expression in E. coli carrying our construct. Bacterial cultures expressing either Met-GS-mRFP or BBa_K880005 (empty vector) were grown overnight at 37°C, lysed and run on SDS-PAGE gels. The expected size of Met-GS-mRFP is approximately 35 kDa, but we observed a strong signal at approximately 36 kDa in the Met-GS-mRFP lysate sample, which was not present in the empty vector sample. This discrepancy in size is likely due to post-translational modifications on the protein such as phosphorylation and glycosylation.

To verify Met-GS-mRFP expression in E. coli, we subjected Met-GS-mRFP lysate to SDS-PAGE, expecting a signal at around 35 kDa. Instead, we saw a signal at around 36 kDa in the Met-GS-mRFP lane, but not in the empty lane that was used as a control. This discrepancy in size is likely due to post-translational modifications on the protein such as phosphorylation and glycosylation. In this gel, Met refers to BBa_K2921500, which contains BBa_1460002 and Met-RFP refers to BBa_K2921520, which contains BBa_1460002.
Functional Assay with Nickel
Our construct produces intracellular Met-GS-mRFP proteins expected to increase the cells’ capacity to store nickel ions. To test the functionality of this protein, we detected the difference in the nickel ion storage capacity of construct-expressing cells and negative-control cells. Thus, our experimental group was cells expressing the Met-GS-mRFP fusion protein. Our negative control group was cells expressing mRFP only. In order to measure cell storage capacity, we incubated cells with the nickel ions over time, to allow the nickel ions to diffuse in and out of the cell. Theoretically, for our experimental groups, the nickel ions would diffuse into the cell and bind to the active site of the intracellular metallothionein protein, reducing the amount of nickel ions diffusing out of the cell. After 2 hours of incubation, we measured the absorbance of nickel ions in the extracellular solution.
By the Beer-lambert law, concentration is directly proportional to absorbance. Thus, for the experimental groups, we expected the extracellular solution to have a lower concentration of nickel ions and, thus, a lower absorbance as compared to the negative control. Nickel solution was prepared by dissolving NiSO4 • 6H2O in distilled water. To optimize the absorbance measurements in the downstream experiment, the wavelength at the peak absorbance of metal solutions were first determined using a spectrophotometer.

Experimental setup: measuring the peak absorbance of nickel solution. NiSO4(H2O)6 was dissolved in distilled water for a 25 mM Ni solution. The solution was measured for its absorbance across the full visible light spectrum using a spectrophotometer.
Overnight bacterial cultures were prepared and standardized to the lowest OD600 across all four groups. Then, the cultures were centrifuged and the pellet was resuspended in nickel solution. The cell-nickel mixtures were gently shaken at room temperature for 2 hours. The cells were then spun down to isolate extracellular solution as the supernatant. The peak absorbance of the nickel ions in the supernatant was measured using a spectrophotometer blanked with distilled water.
Overnight bacterial cultures were prepared and standardized to an OD600 of 0.7. Then, the cultures were centrifuged and the pellet was resuspended in nickel solution. The cell-nickel mixtures were gently shaken at room temperature for 2 hours. The cells were then spun down to isolate extracellular solution as the supernatant. The peak absorbance of the nickel ions in the supernatant was measured using a spectrophotometer blanked with distilled water.

Experimental setup: measuring extracellular concentrations of cell-metal mixtures. The pelleted bacteria were resuspended in nickel solution. After gently shaking the mixture for 2 hours, the absorbance at 651.4 nm of the supernatant was measured using a spectrophotometer. It is expected that the extracellular solution of the experimental group has a lower absorbance than the negative control.
Our results indicate that there are lower absorbance values at the peak absorbance of nickel, 651.4 nm, for cells expressing the Met-GS-mRFP fusion protein, as compared to the RFP only negative control. There is a percent difference of -31.7% between the mean absorbance values of the experimental and control group, suggesting a decrease in extracellular nickel concentration in the presence of Met-GS-mRFP. This shows that proteins are capable of binding to nickel ions, thus increasing the cell’s ability to retain metal ions from their environment.

Met-GS-mRFP increases cellular retention of nickel ions. After two hours of shaking incubation with 25 mM nickel (II) ions, all samples were centrifuged to isolate extracellular solution. At 651.4 nm (the absorbance peak of nickel ions), lower absorbance was observed in the extracellular solution of cells expressing Met-GS-mRFP. Cells expressing RFP only were used as a negative control. Error bars represent standard error. There is a -31.7% percent difference between the mean absorbance values of the experimental and control group, suggesting a decrease in extracellular nickel concentration in the presence of Met.
Color visibility
To verify that the mRFP exhibits red even after being linked to Met, we compared the absorbance at 607nm, which is the emission peak of mRFP (Basic part: BBa_E1010), of cells expressing Met-GS-RFP fusion protein to that of cells expressing the Met protein only (BBa_K2921500, which contains BBa_1460002). First, we standardized overnight bacterial cultures of cells expressing the Met-GS-RFP protein and the Met protein only (BBa_K2921500, which contains BBa_1460002), which should not exhibit color, to the same OD700 to normalize the cell density of the two groups. Then, we diluted the two groups with LB by a ratio of 1:2 to generate a total of four groups for this experiment. After, we used a spectrophotometer to determine the absorbance of the four groups. It is expected that the cells expressing the Met-GS-mRFP protein will have a higher absorbance value at OD607, as it is the peak wavelength of mRFP.

The mRFP in the Met-GS-mRFP fusion protein does exhibit the color pink. The absorbance at 607nm of the cells expressing the Met-GS-mRFP protein is higher than the cells expressing the Met protein only (BBa_K2921500, which contains BBa_1460002), which does not have the fluorescent protein..

The mRFP in the Met-GS-mRFP fusion protein does visibly exhibit the color pink. The overnight liquid culture of cells expressing the Met-GS-mRFP protein is much darker than the cells expressing the Met protein only (BBa_K2921500, which contains BBa_1460002), which does not express the fluorescent protein..
===========================================================
Affinity Test of K1460002
Group: CU 2019
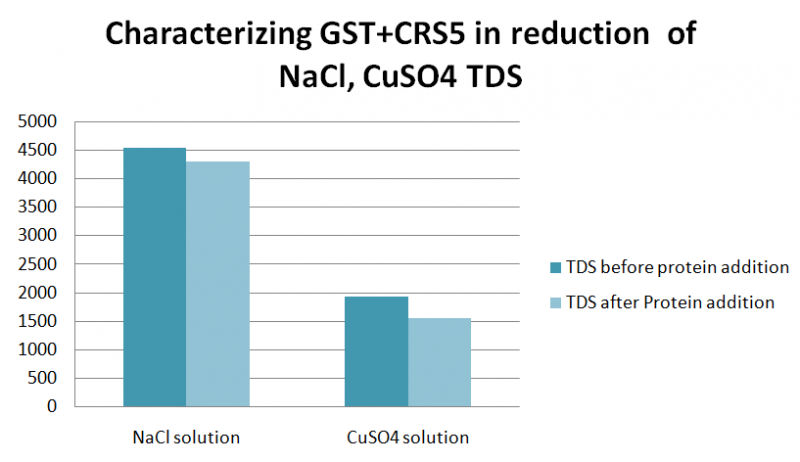
We wanted to test the affinity of the protein for sodium and copper salts. So we performed an experiment where two solutions of NaCl(35g/l) and CuSo4 were prepared and a TDS was measured before and after adding the protein.The GST+CRS5 protein was found to reduce the amount of TDS in NaCl and CuSO4 Solutions; with reduction 288,390 ppm, respectively.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 706
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 118
| None |
