Part:BBa_K1379001
PcomFA
SigmaX dependent promoter initiating the transcription of helicase in Streptococcus pneumoniae NCTC7465 strain. PcomFA (also known as comFAp) is a promoter which proves to be functional in E. coli and S. pneumonia. The sigmaX protein will bind to 8 base pairs of Phelicase promoter and trigger gene expression. The sigmaX gene and Phelicase promoter used in the construct are both cloned from S. pneumoniae NCTC 7465 strain. R.P.U (Relative Promoter Unit) of Phelicase is measured to represent promoter strength in reference to constitutive promoter BBa_J23101. Phelicase promoter is characterized in E. coli DH10B cell as it is frequently used by iGEM participants.
Objective
The objective is to characterize Phelicase promoter so we know whether it iss working in E. coli DH10B strain or not, and to know the R.P.U (Relative Promoter Unit) with BBa_J23101 constitutive promoter as a reference.
Method
By linking Phelicase promoter with GFP generator (BBa_E0240), and SigmaX Generator (BBa_K1379006), the promoter activity was represented by the GFP synthesis rate. Fluorescence was measured when the cells are in the mid-log phase. OD595 values were measured and converted to OD600 to obtain the R.P.U of the promoter.
Characterization
To measure the RPU (Relative Promoter Unit) of Phelicase promoter, we adopted the method described by Kelly et al. in “Measuring the activity of BioBrick promoters using an in vivo reference standard” (Kelly et al., 2009). With the use of EnVision multilabel reader from Perkin Elmer Company, it enabled us to obtain the fluorescence and absorbance of cells over time. GFP intensity and OD595 values were measured every 30 minutes after the E. coli strains are cultured to mid-log phase.
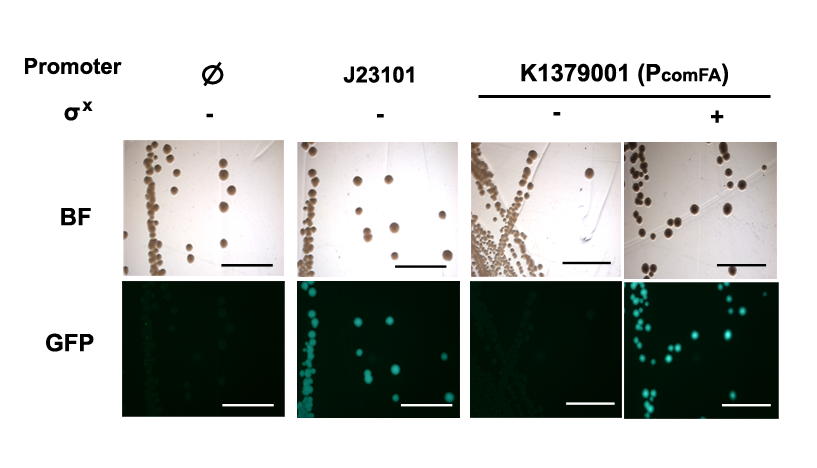
The positive control used in this characterization is BBa_I20260 which is a constitutive promoter BBa_J23101 containing GFP generator BBa_E0240, while the negative control used in this characterization is BBa_K1379003 which is Phelicase promoter with GFP generator but without SigmaX generator.
The detailed description including characterization procedure and Data processing of our characterization can be found in [http://2014.igem.org/Team:Hong_Kong_HKUST/pneumosensor/characterization iGEM HKUST 2014 Wiki Page].
Discussion
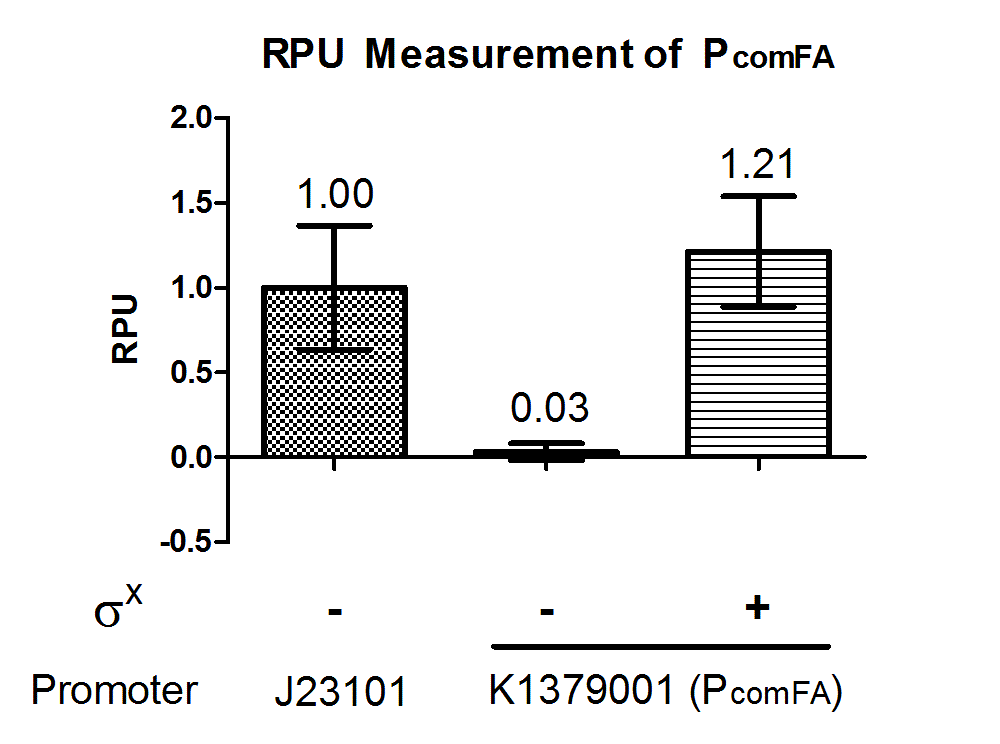
Phelicase promoter is working in E. coli DH10B strain, and express GFP when induced with SigmaX protein. In comparison to the reference promoter BBa_J23101, the R.P.U (Relative Promoter Unit) of PcelA promoter is around 1.2, which is higher than the reference. From Figure 2, we could tell that this promoter is specific as there is almost no GFP expression in the absence of SigmaX protein inducer. This might indicate that the 8 basepairs combox region in Phelicase promoter is specific to SigmaX. However, further characterization needs to be done to evaluate the crosstalk between this inducer-promoter system.
Reference
J. R. Kelly, A. J. Rubin, J. H. Davis, J. Cumbers, M. J. Czar, ..., D. Endy. (2009). Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of Biological Engineering, 3, 4. doi: 10.1186/1754-1611-3-4
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//classic/regulatory/uncategorized
| None |
