Difference between revisions of "Part:BBa K1362401"
(→Nostoc punctiforme DnaE split intein (Npu DnaE)) |
|||
| Line 23: | Line 23: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1362401 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1362401 SequenceAndFeatures</partinfo> | ||
| + | <br> | ||
| + | |||
| + | ===Characterization in <i>Nicotiana benthamiana</i>=== | ||
| + | |||
| + | <b>Group</b>: [http://2016.igem.org/Team:Valencia_UPV Valencia_UPV team 2016] | ||
| + | |||
| + | <b>Author</b>: Valencia UPV iGEM team (Iván Casas Rodrigo, Mónica Victoria Gutiérrez Salazar, Alicia Climent Catalá, Álvaro Ballesteros González) | ||
| + | |||
| + | <b>Summary</b> | ||
| + | |||
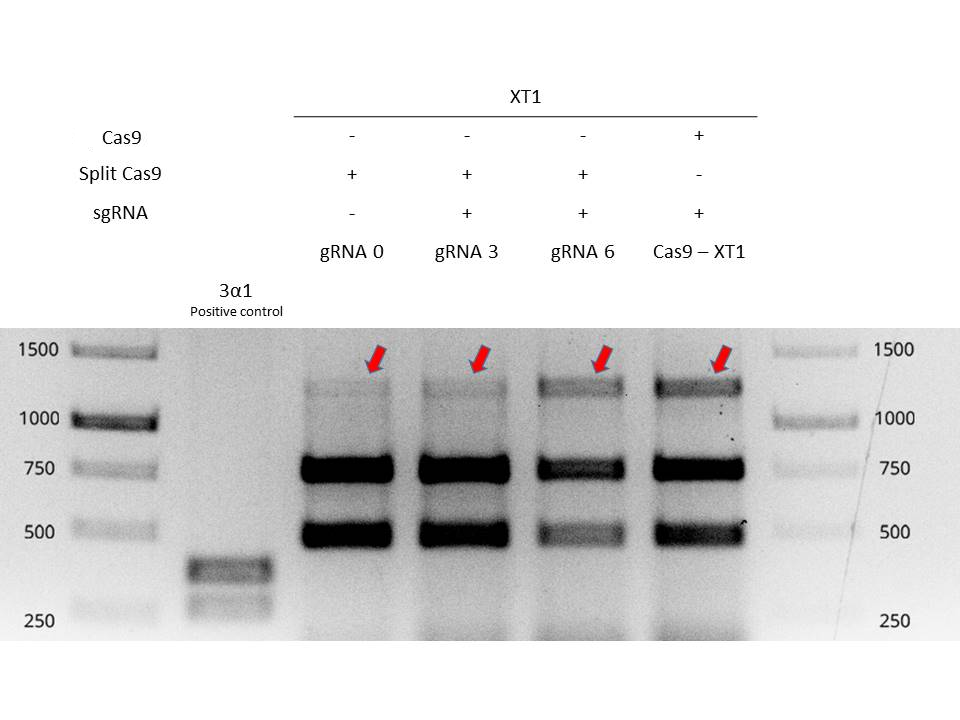
| + | Valencia UPV 2016 team tested and demonstrated the functionality of inteins in a plant chassis, specifically in <i>Nicotiana benthamiana</i>. We used the inteins with our split-Cas9 ([https://parts.igem.org/Part:BBa_K2017000 BBa_K2017000] and [https://parts.igem.org/Part:BBa_K2017001 BBa_K2017001]). The inteins allowed to fusion of the two parts of the split-Cas9 in vivo, in order to obtain a fully functional Cas9 protein. In Figure 1 it is shown an electrophoresis gel with the results of the testing of split-Cas9 in plants. For further information about the experimental design, check the information of split-Cas9 parts, [https://parts.igem.org/Part:BBa_K2017000 BBa_K2017000] and [https://parts.igem.org/Part:BBa_K2017001 BBa_K2017001] | ||
| + | |||
| + | [[Image:BBa_K201700_digestionsplitcas9.jpg|600px|thumb|center|'''Figure 1:''' Electrophoresis gel of XT1 amplicons digestion. Mutation efficiencies of Split-intein-Cas9 system and classical Cas9 system are compared by resistant bands signal intensity.]] | ||
Revision as of 14:53, 22 October 2016
NpuDnaE C-Intein cloning piece
Nostoc punctiforme DnaE split Intein C-terminal half. This is a DNA piece for cloning used to assemble our other BioBrick parts.
Nostoc punctiforme DnaE split intein (Npu DnaE)
Inteins can be found in all kingdoms of life, however their use for the organism remains yet unknown. In the past years many intein sequences were found in the genomes of cyanobacteria. Npu DnaE is a naturally split intein from the alpha subunit of Dna Polymerase III in Nostoc punctiforme bacteria which shows [1] extraordinarly high splicing activity and therefore is currently still on of the gold-standards for inteins.
Amino Acid Sequence: N-Intein: AEY | CLSYETEILTVEYGLLPIGKIVEKRIECTVYSVDNNGNIYTQPVAQWHDRGEQEVFEYCLEDGSLIRATKDHKFMTVDGQMLPIDEIFERELDLMRVDNLPN C-Intein: MIKIATRKYLGKQNVYDIGVERDHNFALKNGFIASN | CFN
The sequences above are specified with three native extein residues on each of the termini. Please note that in the case of Npu DnaE these are not the most efficient amino acids in supporting the splicing reaction. Detailed information that we found to be very useful is given in [2].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization in Nicotiana benthamiana
Group: [http://2016.igem.org/Team:Valencia_UPV Valencia_UPV team 2016]
Author: Valencia UPV iGEM team (Iván Casas Rodrigo, Mónica Victoria Gutiérrez Salazar, Alicia Climent Catalá, Álvaro Ballesteros González)
Summary
Valencia UPV 2016 team tested and demonstrated the functionality of inteins in a plant chassis, specifically in Nicotiana benthamiana. We used the inteins with our split-Cas9 (BBa_K2017000 and BBa_K2017001). The inteins allowed to fusion of the two parts of the split-Cas9 in vivo, in order to obtain a fully functional Cas9 protein. In Figure 1 it is shown an electrophoresis gel with the results of the testing of split-Cas9 in plants. For further information about the experimental design, check the information of split-Cas9 parts, BBa_K2017000 and BBa_K2017001