Difference between revisions of "Part:BBa K112808:Experience"
(→Platereader Tests of Lysis) |
|||
| Line 4: | Line 4: | ||
===Applications of BBa_K112808=== | ===Applications of BBa_K112808=== | ||
| + | |||
| + | |||
| + | == Using the Lysis Cassette BBa_K112808 To Recover DNA == | ||
| + | |||
| + | === Introduction === | ||
| + | |||
| + | The 2011 EPFL iGEM team setup an information processing system that recovers DNA from cells in which a strong binding affinity exists between a particular transcription factor and its promoter. More specifically, our system looks at the interaction between the TetR transcription factor (TF) and its promoter pTet. If the interaction between these two is strong, the system will induce lysis in the cell. As a result of the lysis, the supernatant will contain the plasmids with the sequences for the transcription factor and the promoter. The information processing part comes in when one is interested in finding mutant transcription factor sequences and/or mutant promoter sequences with weaker or stronger binding affinities than the wildtype. In such a case, our system is ideal since a whole culture of cells -- each containing different plasmids with random combinations of mutant TFs and mutant promoters -- can be grown and the lysis will select the cells whose binding affinity was strongest -- a "survival of the weakest" strategy. One need only sample the supernatant and run through the system a few more times to select the best binding sequences of your favorite transcription factor and promotor. | ||
| + | |||
| + | === Platereader Tests of Lysis === | ||
| + | |||
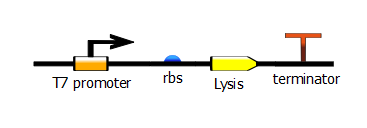
| + | To test this selection system, we enhanced the current BBa_K112808 lysis cassette by adding a T7 promoter upstream of the cassette. To find out more about this construct, please refer to the registry page that presents more details. | ||
| + | |||
| + | |||
| + | [[File:T7_rbs_lysis_term.png]] | ||
| + | |||
| + | === Experiment Details === | ||
| + | |||
| + | 1) We made liquid cultures of BL21 cells containing the plasmids with the lysis cassette driven by a T7 promoter. These plasmids have Kanamycin resistance, so the cultures were made using 5 mL of LB medium with 5 uL of 100 ug/uL Kanamycin and grown overnight. | ||
| + | |||
| + | 2) We made 1:10 dilutions of cells to LB medium by adding 100 uL of cells to 900 uL of LB with 1 uL of Kanamycin. From this 1 mL mix, we pipetted 100 uL into the wells of a 96 well plate. | ||
| + | |||
| + | 3) We covered each well with 20 uL of mineral oil to avoid evaporation. Then we set the platereader to shake continuously and take optical density measurements at 600 nm wavelength every ten minutes. | ||
| + | |||
| + | 4) We waited an hour and a half (approximately) until the cells had reached their log-linear growth phase. Then we took out the plate and added 1 uL of IPTG to each well. We made triplicate data for each concentration of IPTG (0 uL for negative control, 50 uL, 100 uL, 250 uL, and 500 uL). | ||
| + | |||
| + | 5) We set the plate again into the platereader, ran it for 12 hours with continous shaking. Optical density measurements were made every ten minutes. | ||
| + | |||
| + | === Live Culture Tests of DNA Recovery === | ||
| + | |||
====BBa_K112808 - Enterobacteria phage T4 Lysis Device - UNIPV-Pavia Team (test performed by L. Pasotti, M. Lupotto, S. Zucca, R. De Molfetta)==== | ====BBa_K112808 - Enterobacteria phage T4 Lysis Device - UNIPV-Pavia Team (test performed by L. Pasotti, M. Lupotto, S. Zucca, R. De Molfetta)==== | ||
| Line 116: | Line 145: | ||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
<!-- DON'T DELETE --><partinfo>BBa_K112808 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K112808 EndReviews</partinfo> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 20:33, 20 September 2011
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K112808
Using the Lysis Cassette BBa_K112808 To Recover DNA
Introduction
The 2011 EPFL iGEM team setup an information processing system that recovers DNA from cells in which a strong binding affinity exists between a particular transcription factor and its promoter. More specifically, our system looks at the interaction between the TetR transcription factor (TF) and its promoter pTet. If the interaction between these two is strong, the system will induce lysis in the cell. As a result of the lysis, the supernatant will contain the plasmids with the sequences for the transcription factor and the promoter. The information processing part comes in when one is interested in finding mutant transcription factor sequences and/or mutant promoter sequences with weaker or stronger binding affinities than the wildtype. In such a case, our system is ideal since a whole culture of cells -- each containing different plasmids with random combinations of mutant TFs and mutant promoters -- can be grown and the lysis will select the cells whose binding affinity was strongest -- a "survival of the weakest" strategy. One need only sample the supernatant and run through the system a few more times to select the best binding sequences of your favorite transcription factor and promotor.
Platereader Tests of Lysis
To test this selection system, we enhanced the current BBa_K112808 lysis cassette by adding a T7 promoter upstream of the cassette. To find out more about this construct, please refer to the registry page that presents more details.
Experiment Details
1) We made liquid cultures of BL21 cells containing the plasmids with the lysis cassette driven by a T7 promoter. These plasmids have Kanamycin resistance, so the cultures were made using 5 mL of LB medium with 5 uL of 100 ug/uL Kanamycin and grown overnight.
2) We made 1:10 dilutions of cells to LB medium by adding 100 uL of cells to 900 uL of LB with 1 uL of Kanamycin. From this 1 mL mix, we pipetted 100 uL into the wells of a 96 well plate.
3) We covered each well with 20 uL of mineral oil to avoid evaporation. Then we set the platereader to shake continuously and take optical density measurements at 600 nm wavelength every ten minutes.
4) We waited an hour and a half (approximately) until the cells had reached their log-linear growth phase. Then we took out the plate and added 1 uL of IPTG to each well. We made triplicate data for each concentration of IPTG (0 uL for negative control, 50 uL, 100 uL, 250 uL, and 500 uL).
5) We set the plate again into the platereader, ran it for 12 hours with continous shaking. Optical density measurements were made every ten minutes.
Live Culture Tests of DNA Recovery
BBa_K112808 - Enterobacteria phage T4 Lysis Device - UNIPV-Pavia Team (test performed by L. Pasotti, M. Lupotto, S. Zucca, R. De Molfetta)
Description
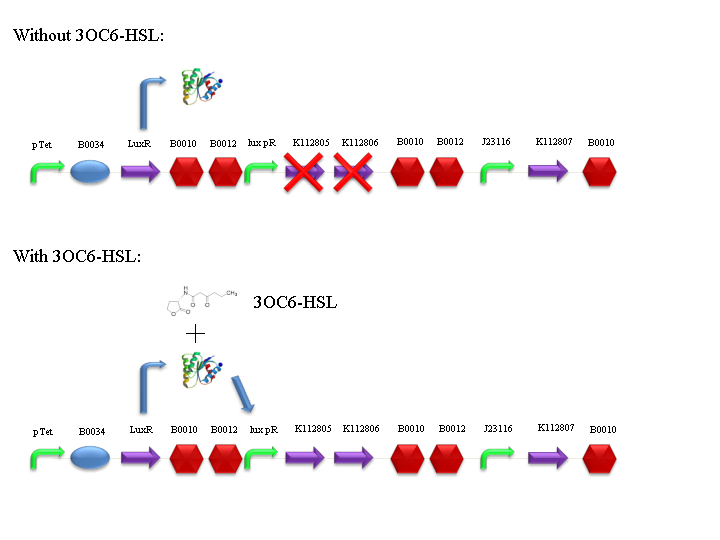
The UNIPV-Pavia Team decided to build up a specific measurement system: BBa_K173015, in which we can modulate the expression of the lysis genes through BBa_F2620 inducible device. During this summer we have also characterized BBa_F2620 in standard units, so the characterization of the lysis device is promoter-independent.
BBa_F2620 receiver device controls the expression of holin (BBa_K112805) and lysozyme (BBa_K112806) genes from T4 bacteriophage. Holins form pores in the inner membrane of bacteria, while lysozymes degrade the peptidoglycan layer, passing through these pores and performing the lysis of the bacterial cell. This device also contains a constitutive expression of an antiholin (BBa_K112807), which inhibits holin multimer formation.
Lysis can be induced with 3OC6-HSL autoinducer molecule: it binds luxR protein (encoded by BBa_C0062), which is constitutively expressed by tetR promoter (BBa_R0040). LuxR-HSL complex can work as a transcriptional activator for lux promoter (BBa_R0062).
Characterization
Compatibility: E. coli TOP10 in pSB1A2.
We performed four experiments on our measurement system (BBa_K173015) in order to characterize BBa_K112808 activity.
All the experiments have been performed in the same conditions in the microplate reader and involved the induction of BBa_K173015 with 3OC6-HSL 10 nM (which corresponds to a RPU value of 4.44 [3.81 ; 5.44] in LB for BBa_F2620) and 100 nM (which corresponds to a RPU value of 4.53 [3.88 ; 5.18] in LB for BBa_F2620). The basic activity of BBa_F2620 was ~0 RPU.
O.D.600 has been used to measure the amount of bacteria in the wells.
Experiment 1 results
We tested BBa_K173015 and BBa_K173010 as a negative control. These two parts were tested according to Microplate reader experiments section, except for BBa_K173015 first inoculum, which was performed from single colonies in a streaked LB agar plate. Three colonies were tested separately for this part.
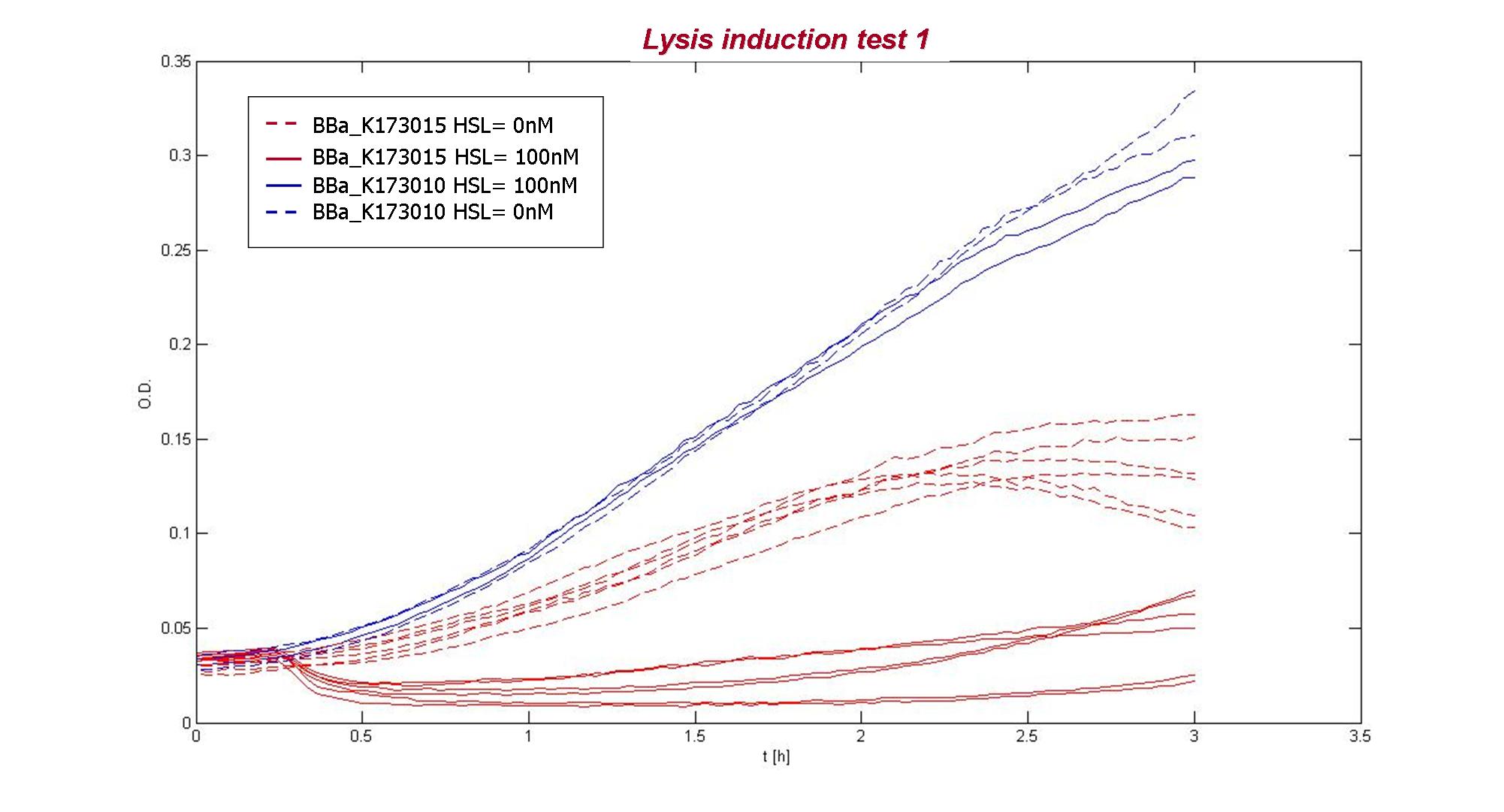 O.D.600 of two replicates of three colonies of BBa_K173015 (uninduced or induced with 100 nM 3OC6-HSL) and of BBa_K173010. |
The result showed that BBa_K173010 had comparable growth curves in presence of 3OC6HSL (dashed blue lines) or not (blue lines). The cultures containing the lysis device not induced (dashed red lines) shows a slow growth compared to BBa_K173010, maybe because of leakage of the inducible system causing a weak lysis or because the device decrease bacteria’s fitness. The culture containing the induced lysis device (red lines) shows a rapid decrease of O.D. after 15 minutes from induction, followed by a O.D. increasing. Different colonies had comparable dynamic growth.
This experiment has been repeated two times (data have not been reported): the first one (Experiment 2) lysis failed for unknown reasons, while in the second one (Experiment 3) it worked as in Experiment 1.
We deduced that the slow O.D. increase in lysed cultures (as well as the apparent genetic instability of the inducible device, which was unresponsive to 3OC6-HSL in Experiment 2) could be due to a positive selection of mutant cells unresponsive to 3OC6-HSL (or defective in the lysis device) or to the shortage of free 3OC6-HSL. To test this hypotheses, we performed a fourth, independent experiment, using BBa_K173015 inducible system, BBa_B0033 as a negative control and a culture containing only BBa_K112808 part (not functional because the operon lacks a promoter). The test has been performed with two different inducer concentrations (100 nM and 10nM) and following the standard protocol of Microplate reader experiments. In this experiment (Experiment 4), lysis was induced at t=0 and, in previously induced cultures, also after 4 hours of growth to test if they were still responsive to 3OC6-HSL.
 O.D.600 of BBa_K173015 (uninduced, induced with 10 nM 3OC6-HSL or 100 nM), of BBa_K173010 and of BBa_K112808. |
The growth curves of the second experiment are in agreement with the result of Experiment 1. Moreover, our hypothesis of mutant generation for BBa_K173015 induced culture is confirmed by the second induction: if the increase in O.D. was perhaps due to a shortage of 3OC6-HSL in the growing population, the second induction would have caused a new decline in O.D., which is not our case. This hypotesis is also confirmed by the growth curves of induced (at t=0) BBa_K173015 cultures, which show a decrease (from 15 min to 2 hours) followed by an increasing growth, comparable with the negative controls growth and faster than BBa_K173015 non induced cultures growth.
This new experiment also shows that both induction with 10nM or 100nM of 3OC6-HSL cause the same decrease in O.D. value and that K112808 part alone (green lines) could grow like the negative control, demonstrating that the slow growth of BBa_K173015 was actually due to leakage activity of BBa_F2620.
Conclusions
We built up a new measurement system to characterize a particularly interesting lysis device.
The reported results show that the lysis device works as expected when gene expression of the lysis genes is triggered. The performed experiments showed a genetic instability of this device (which was expected because a complete expression system of lysis genes is introduced in E. coli): in one of four experiments lysis could not be induced (Experiment 2) and in all the other experiments after lysis induction in BBa_K173015 cultures showed an O.D.600 decrease, followed by a fast increase, probably due to mutations in lysis expression system.
Growth conditions
Microplate reader experiments
- 8 ul of long term storage glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours.
- The grown cultures were then diluted 1:100 in 5 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours.
- These new cultures were diluted to an O.D.600 of 0.02 (measured with a TECAN F200 microplate reader on a 200 ul of volume per well; it is not comparable with the 1 cm pathlength cuvette) in a sufficient amount of medium to fill all the desired microplate wells.
- These new dilutions were aliquoted in a flat-bottom 96-well microplate, avoiding to perform dynamic experiments in the microplate frame (see [http://2009.igem.org/Team:UNIPV-Pavia/Methods_Materials/Evaporation Frame effect section] for details). All the wells were filled with a 200 ul volume.
- If required, 2 ul of inducer were added to each single well.
- The microplate was incubated in the Tecan Infinite F200 microplate reader and fluorescence (when required) and absorbance were measured with this automatic protocol:
- 37°C constant for all the experiment;
- sampling time of 5 minutes;
- fluorescence gain of 50;
- O.D. filter was 600 nm;
- GFP filters were 485nm (ex) / 540nm (em);
- 15 seconds of linear shaking (3mm amplitude) followed by 10 seconds of waiting before the measurements in order to make a homogeneous culture.
- Variable experiment duration time (from 3 to 24 hours).
Materials
- Long term glycerol stocks were stored at -80°C with a final glycerol concentration of 20%
- Antibiotics were: Ampicillin (Amp) 100 ug/ml. It was stored at -20°C in 1000x stocks. Amp was dissolved in water.
- LB medium was prepare with: 1% NaCl, 1% bactotryptone, 0.5% yeast extract. The medium was not buffered with NaOH.
- 3OC6-HSL (Sigma) was dissolved in water and stored at -20°C in a 2mM stock.
User Reviews
Sequencing analysis of iGEM QC 2009 performed by UNIPV-Pavia team 2009
After the alignment of this BioBrick sequence from 2009 iGEM HQ quality contol with the theoretical sequence, many difformities have been reported (sequencing results have been taken from iGEM QC):
- the alignment showed several gaps in the scars;
- there is a silent point mutation in holin gene;
- the transcriptional terminator at the end of the sequence is actually K112710, not B0010.
However, the part should work properly because the regulatory parts and the amino acid sequence of the genes are correct. This has also been confirmed by functional tests (see Applications section).
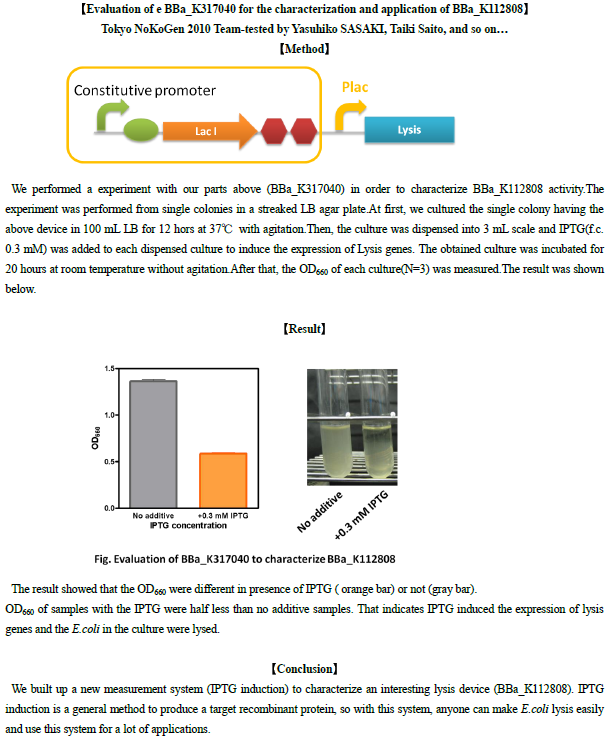
Evaluation of e BBa_K317040 for the characterization and application of BBa_K112808
UNIQcf727d37357b157b-partinfo-00000020-QINU UNIQcf727d37357b157b-partinfo-00000021-QINU