Part:BBa_I712667
HIV-1 aspartyl protease
HIV-1 protease is an aspartyl protease that is essential for the life-cycle of HIV and cleaves proteins at specific aminoacid sequence.
Contribution: XHD-Wuhan-Pro-China 2021
Literature 1 introduction
One important element in the life cycle of HIV retrovirus that researchers target at to develop inhibitors is HIV-1 protease (HIV PR), an essential enzyme needed in the proper assembly and maturation of infectious virions. Understanding the chemical mechanism of this enzyme has been a basic requirement in the development of efficient inhibitors. In this review, we summarize studies conducted in the last two decades on the mechanism of HIV PR.
The role of HIV PR in the life cycle of HIV
After the host cell is activated, transcription of the viral DNA into messenger RNA occurs. The messenger RNA is then translated into viral proteins. HIV protease is required in this step to cleave a viral polyprotein precursor into individual mature proteins.
Mechanism of the HIV protease
According to several studies, HIV PR in general has been shown to belong to the class of the aspartic proteases. Examining the sequence homology of HIV PR to other cellular aspartic proteases shows that this enzyme has the sequence Asp-Thr-Gly, which is conserved among the aspartic protease enzymes. These results suggested that HIV PR-1 enzyme may have similar structural features to the aspartic protease enzymes as well as a similar mechanism.
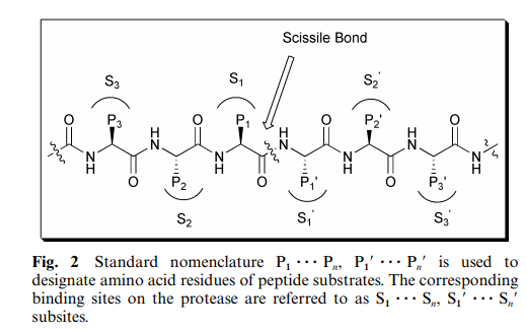
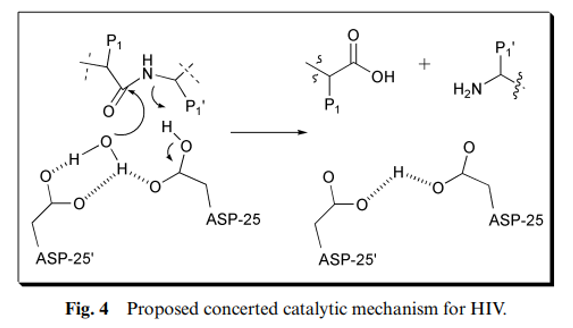
Although the HIV PR mechanism shares many features with the rest of the aspartic protease family, the full detailed mechanism of this enzyme remains not fully understood. Jaskólski et al. have proposed a new model of the enzymatic mechanism for the HIV PR enzyme based on the crystal structure of a complex between a chemically synthesized HIV PR and an octapeptide inhibitor U-85548e (H-Val-Ser-Gln-Asn-Leu-ψ-[CH(OH)-CH 2 ]-Val-Ile-Val-OH), as well as other crystal structures of HIV PR complexed with different inhibitors. In this mechanism (Fig. 4), the hydrolysis reaction is viewed as a one-step process during which the nucleophilic water molecule and the acidic proton attack the scissile peptide bond in a concerted manner. The issue of the simultaneous attack from the nucleophile and electrophile is the major difference between this mechanism and other previously proposed mechanisms.
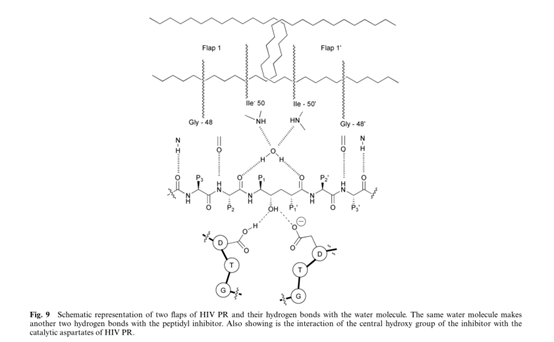
In HIV PR there are two flaps in the active dimer compared to the single flap in non-viral aspartic protease. 35 Moreover, while in the pepsin-like protease there is a direct H-bonding between the flap and the substrate, in HIV PR the carbonyl group between P1 and P2 and the carbonyl between P1’ and P2’ both make hydrogen bonds to the same water molecule (Fig. 9).This water molecule makes another two hydrogen bonds to the flaps to create approximately tetrahedral geometry. The presence of this water molecule has been observed in most of the crystal structures of HIV PR bound to different inhibitors. It has been proposed that this water molecule is relevant for catalysis and a key feature in the differences of the mechanisms of the retroviral HIV PR and the corresponding cell-encoded enzymes. The hydrogen bonds of this water molecule apply strain on the scissile amide bond, causing it to rotate out of the plane and lose its double bond character, which enhances its vulnerability towards hydrolysis. Using total chemical synthesis of proteins, HIV PR was synthesized in which the Gly49 –Ile 50 N(H)-atom (Fig. 9) was specifically replaced by an O-atom, thus deleting one of the hydrogen bonds from one of the flaps to the water molecule. The resulting enzyme with the single flap–substrate hydrogen bonds was fully active. Based on these results, it has been suggested that this enzyme may make use of only one flap in the catalysis, showing a similarity to pepsin-like enzymes.
Mechanism based strategy for drug design
The Dupont Merck group has designed a new generation of HIV PR inhibitors based on cyclic urea cores. The uniqueness of this class of inhibitors is the cyclic urea carbonyl oxygen that mimics the hydrogen bonding features of the key structural water molecule discussed earlier (Fig. 9). The group reasoned that incorporating the cyclic urea core into an inhibitor would replace the flap water molecule, which could lead to better binding energy due to the positive entropic effect that should be provided by the cyclic urea core. These inhibitors also contained the diol functionality as a transition state mimic to interact with the catalytic aspartates.
Conclusion
It is clear from all the previous studies that many researchers agree about the similarity of the mechanism to that of the other aspartic proteases: the involvement of the water molecule as nucleophile, the general role of the two active site aspartyl residues.
Reference
Brik, A. and Wong, C., 2003. HIV-1 Protease: Mechanism and Drug Discovery. ChemInform, 34(18).
Literature 2 introduction
In order to answer the question of how the HIV-1 aspartyl protease actually process PrSSgag and Pr160gag-Pol polyproteins contained in the host cell with an immature structure and composition, we undertook a series of genetic studies in Escherichia coli on the maturation of the protease from its precursor forms.
Result
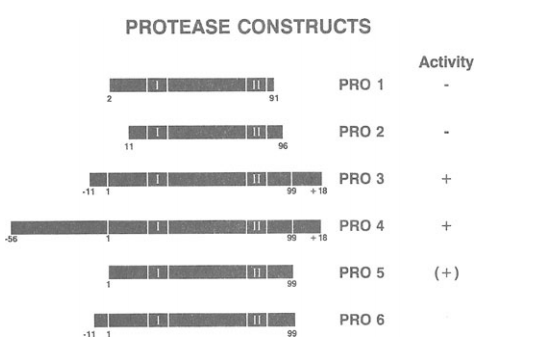
The mature, 99 amino acid-long HIV-1 protease has been expressed in bacteria by several laboratories using genetic constructs that encode the 99-mer species itself or, more typically, a precursor species that is readily processed to the mature form of the enzyme (see 'Viral Proteinases as Targets for Chemotherapy' (1989) as a source of information and references). As a rule, protease with the best attributes of solubility, homogeneity and activity has been obtained from those expression vectors that direct the production of the mature, dimeric protease from a precursor form. Examples of protease constructs that were generated in our laboratory are shown in Figure 1 and are described hereafter.
In our laboratory, the protease expression vectors that yielded the mature HIV-1 protease in its most soluble, homogenous and active form were the ones encoding the PR03 and PR04 constructs. In these constructs, the protease was expressed via 'auto'-processing (probably an intermolecular event) of a larger precursor containing both upstream sequences (11 or S6 residues for PR03 and PR04, respectively) and downstream sequences (18 residues for both PR03 and PR04) from the pol open reading frame. High levels of protease activity were readily demonstrated for the PR03 and PR04 constructs both within whole bacterial cells using a co-expressed PrSSgag polyprotein substrate (Debouck et aI, 1987) and in vitro after purification of the enzyme (Meek et aI, 1989; Strickler et aI, 1989). We also constructed an expression vector, PROS, for the direct production of the mature 99 amino acid protease. For this purpose, a translation initiation codon (ATG) was placed immediately before the first codon of the protease (encoding proline 1) and a stop codon (TAG) immediately following the last codon (encoding phenylalanine 99). This protease construct was found to be active when tested in whole bacterial cells for the specific processing of a co-expressed PrSSgag substrate.
Schematic representation of HIV-1 protease genetic constructs. The HIV-1 protease coding region present in six bacterial expression vectors (PR01 to PR06) is depicted. PR01-PR04 have been previously described (Debouck et aI, 1987). PROS and PR06 are described in the text. The boxed regions marked I and II correspond to the two dOlllains that are highly conserved among retroviral, lIlicrobial and cellular aspartyl proteases (Pearl and Taylor, 1987). The numbering starts with 1 at the first residue (proline) and 99 at the last residue (phenylalanine) of the mature HIV-1 protease;-l1 and -56 indicate the nUlllber of amino acid residues from the pol region upstrealll of the lIlature protease; +18 indicates the nUlllber of residues frolll the pol region downstrealll of the mature protease. The activity of the protease products against PrSsgag in E . coli (co-expression systelll) is shown: +, active, PrSS is processed; - inactive, PrSS is not processed.
Reference
Debouck C., Deckman I.C., Grant S.K., Craig R.J., Moore M.L. (1990) The HIV-1 Aspartyl Protease: Maturation and Substrate Specificity. In: Pearl L.H. (eds) Retroviral Proteases. Palgrave, London. https://doi.org/10.1007/978-1-349-11907-3_3
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |

 1 Registry Star
1 Registry Star