Part:BBa_C0079
lasR activator from P. aeruginosa PAO1(+LVA)
coding region for lasR protein, which accepts chemical signal AI-1 (made by lasI protein)
I. Characterisation of LasR by Imperial College London iGEM 2016
Group: Imperial College London 2016
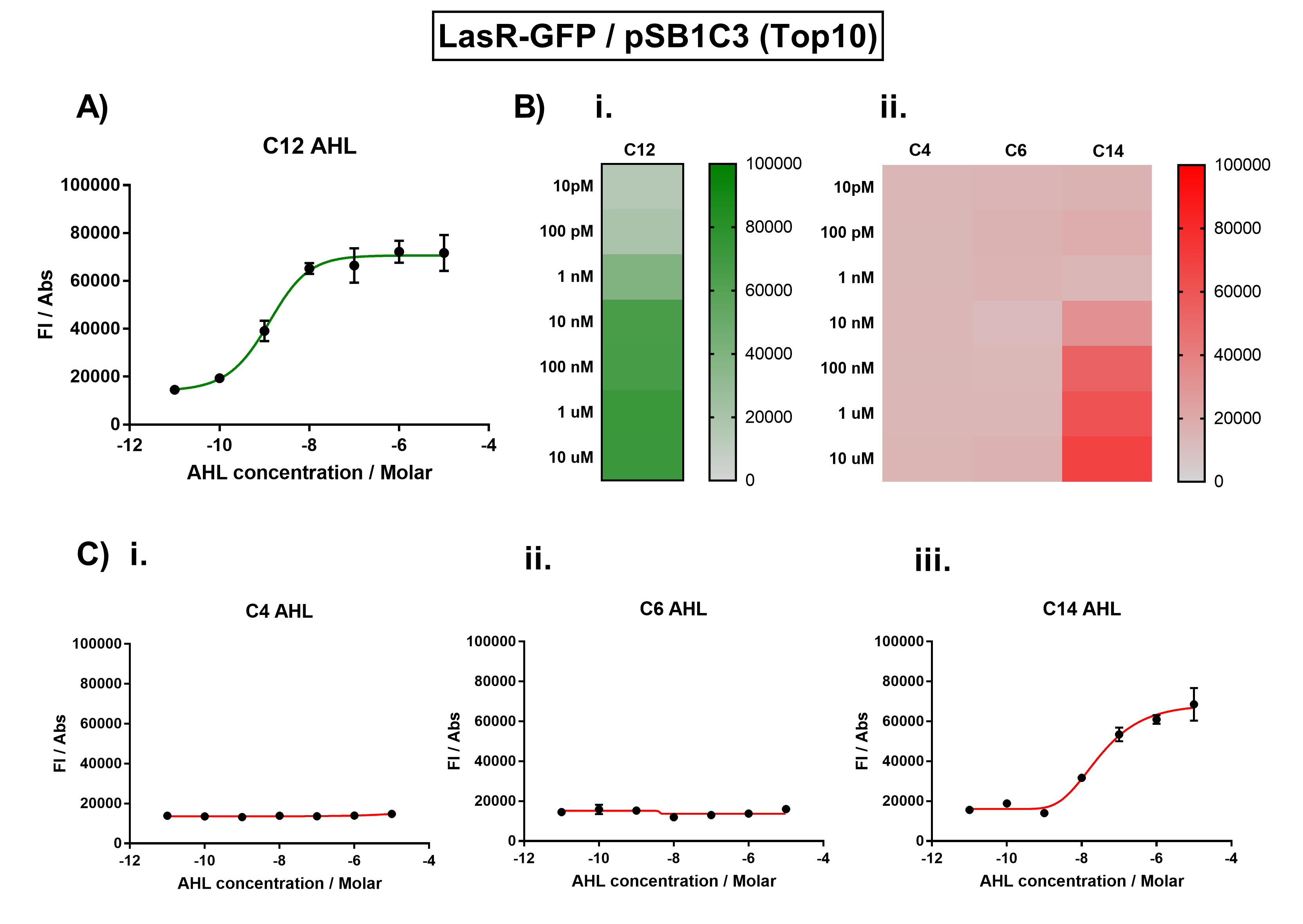
We improved this part by characterising the activation range of LasR, and the crosstalk between LasR and non-cognate AHLs. We placed the coding sequence downstream from the pLas promoter, and recorded the fluorescence for different concentrations of AHLs.
Characterisation data
The activation range of LasR by its cognate inducer (3O-C12-AHL) was characterised in TOP10 E.Coli cells. Cells transformed with the Las response device were cultured to the exponential phase and treated with appropriate concentrations of 3O-C12-AHL in 96-well microplates. These induced cells were grown in the microplates and their fluorescence and absorbance values (OD 600) were monitored over time using a microplate reader. The reported values for the normalised fluorescence represents the values recorded 180 minutes after AHL induction. The normalised fluorescence was calculated by dividing fluorescence values by absorbance values and correcting for LB autofluorescence. In order to characterise the orthogonality of the Las system, we measured absorbance and fluorescence of the cells in the plate reader after treating them with varying concentrations of 3 different AHL signals (C4 AHL of the Rhl system, 3O-C6 AHL of the Lux system, and 3O-C14 AHL of the Cin system). This allowed us to determine whether these non-cognate AHLs were capable of activating the LasR response protein, and therefore the level of crosstalk between the quorum sensing systems.
Figure 1. Characterisation of the Las response device (BBa_K1893001). (A) Transfer function curve of normalised fluorescence against cognate inducer C12-AHL (3O-C12 AHL) concentrations. (B) Heat map of normalised fluorescence of RhlR-GFP system over a range of AHL concentrations: (i) Binding of RhlR-GFP to its cognate AHL (3O-C12 AHL). (ii) Binding of RhlR-GFP to 3 non-cognate AHLs (C4 AHL, 3O-C6 AHL, 3OH-C14 AHL). (C) Transfer function curves of normalised fluorescence against non-cognate inducer AHL (3O-C12 AHL) concentrations to investigate inducer AHL crosstalk: (i) C4-AHL of the Rhl system (ii) C6-AHL (3O-C6 AHL) of the Lux system (iii) C14-AHL (3O-C14 AHL) of the Cin system. Experiments were performed in E. coli Top10 cell strain cultured at 37°C. Normalised fluorescence was calculated by dividing fluorescent signal by cell density (OD600). Fluorescence measurements were recorded at 180 minutes. Reported values represent the mean normalised fluorescence value from 3 technical repeats and error bars represent standard deviation of these.
Analysis The results from Figure 1A show that the concentration range of AHL (3O-C12 AHL) required for the activation of the Las response device was 100pM-10uM. Furthermore, it can be seen from Figure 1C that LasR does not appear to be activated by C4 AHL or 3O-C6 AHL, suggesting that the Las quorum system is orthogonal with Rhl and Lux. However, it is seen that LasR is activated significantly by 3O-C14 AHL, suggesting that the Las and Cin quorum systems lack orthogonality.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 322
Illegal AgeI site found at 519 - 1000COMPATIBLE WITH RFC[1000]
Allergen characterization of BBa_C0079 : Potential Allergen - 60% identity to minor allergen Cla 7 from Davidiella tassiana
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick part, BBa_C0079, there was a 60.0% of identity match and 80% of similarity match to minor allergen Cla 7 [Davidiella tassiana] https://www.ncbi.nlm.nih.gov/protein/467629. This means that the biobrick part HAS Potential Allergen Status. In the 80 amino acid alignments by FASTA, matches found that there are greater than 35% for this biobrick.
//cds/transcriptionalregulator/activator
//function/cellsignalling
//function/regulation/transcriptional
| direction | Forward |
| function | Rep |
| ligands | PAI |
| protein | lasR-LVA |
| swisspro | P25084 |
| tag | LVA |

 1 Registry Star
1 Registry Star