Part:BBa_K3151009
Cyclic-di-GMP Riboswitch
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 16
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
The regulation of eGFP expression is dependent on the interactions between the terminator stem loop region of the riboswitch and the activation or inactivation of this terminator stem loop by the presence or absence of cyclic-di-GMP. When cyclic-di-GMP is bound to the aptamer region of the riboswitch, it forms the terminator loop structure which prevents the expression of eGFP through truncation of the emerging transcripts. From this, we hypothesized that the riboswitch suppresses the expression of green fluorescent protein in the presence of cyclic-di-GMP, hence, this part will express less eGFP than the riboswitch without the terminator stem loop [BBa_K3151002].
Characterisation
In order to test the function of our riboswitch [BBa_K3151009], we developed two riboswitch/eGFP operon constructs; one containing the terminator loop region within the riboswitch and one without. We hypothesised that the construct containing the terminator stem structure would constitutively express eGFP, regardless of cyclic-di-GMP concentration. Because of this constant activity, we refer to this construct as “R ON” (for riboswitch ON) [BBa_K3151002]. Alternatively, the construct with the terminator stem would have the expression of eGFP be repressed by native cellular concentrations of cyclic-di-GMP, with this construct being referred to as “R OFF” (for riboswitch OFF) [BBa_K3151011].
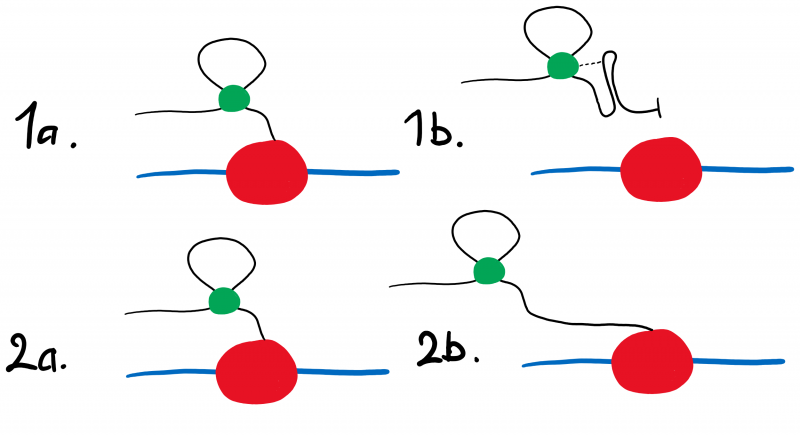
Figure 1: The hypothesised activity of the two riboswitch constructs. While the RNA polymerase (red) moves along the operon (blue), cyclic-di-GMP (green) will bind to the aptamer region of the riboswitch transcript (black). When bound to cyclic-di-GMP, the R OFF construct (Fig. 1.1a) will produce the terminator loop structure (Fig. 1.1b) and cease transcription, preventing the expression of eGFP. When cyclic-di-GMP binds to the R ON transcript (Fig. 1.2a) it does not form the terminator loop structure (Fig 1.2b), thereby having no effect on the expression of eGFP.
We compared the expression of our cyclic-di-GMP riboswitch + eGFP bioparts over 60 hours of incubation between the stationary phase, a Lac (BBa_R0010), and a Tac [BBa_K180000] promoter. All three promoters showed significant increase in expression starting at the stationary phase, however the Lac and Tac promoters showed a linear increase from that point while the Full-length stationary phase osmY promoter significantly increased expression then plateued after several hours.
In order to characterise the mode of action of the riboswitch/promoter combinations, we collaborated with Team Sydney Australia to perform GFP fluorescence assays[1]. These assays were used to quantify the production of eGFP between our reporter constructs, as well as comparing the function of the stationary phase promoter against two inducible promoters, Lac [BBa_R0010] and Tac [BBa_K180000]. Samples were measured with the BMG Pherastar plate reader to measure eGFP (Ex485 nm Em520 nm) and OD (600 nm).
These results demonstrate that while the production of eGFP increases at 6-8 hours for all the promoters (i.e. roughly the point at which samples would be entering the stationary phase), the Lac (Fig. 2 & 5) and Tac (Fig. 3 & 6) inducible promoters show a steady, somewhat linear accumulation of eGFP over time, the stationary phase promoter (Fig. 4) shows a sharp increase and beings to level out after ~20 hours.
These results also demonstrate that the Lac and Tac promoters have significantly higher background expression of the R OFF construct. This is likely due to the strength of the promoters, and the length of the transcripts produced by either construct. This is demonstrated clearly by the results of the Tac promoter (Fig. 3 & 6), in which by the end of the assay the R OFF samples were producing higher levels of eGFP. We believe that this is due to the fact that the binding of cyclic-di-GMP to the transcript results in shorter transcripts, allowing the RNA polymerase to produce more of the transcripts quickly and saturating the cyclic-di-GMP, resulting in a loss of inhibition of eGFP transcription. This effect can also be observed in the samples under control of the Lac promoter (Fig. 2 & 5). By contrast, the background level of transcription under the stationary phase promoter remains constant throughout the entire assay (Fig. 4).
These results are consistent with previous characterisations of the stationary phase promoter, which observed significant increases in β-galactosidase concentration after entry into the stationary phase at 5 hours[2].
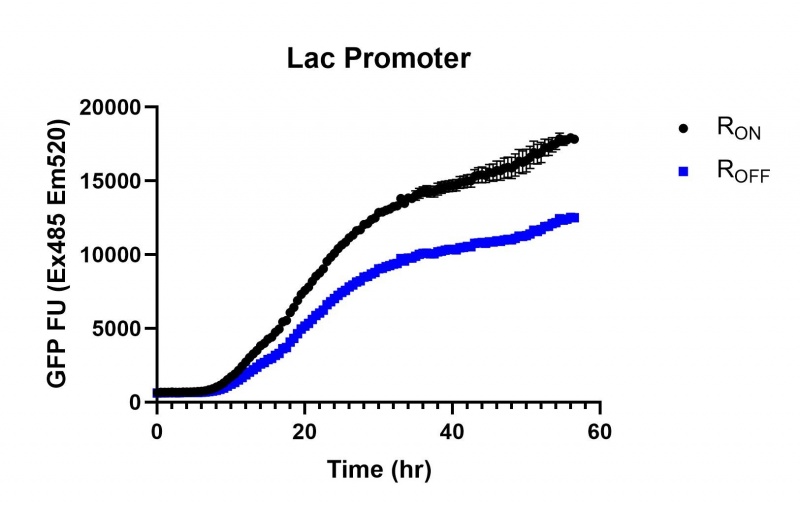
Figure 2: eGFP produced by cells transformed with the Lac riboswitch constructs [BBa_K3151028] over 60 hours.
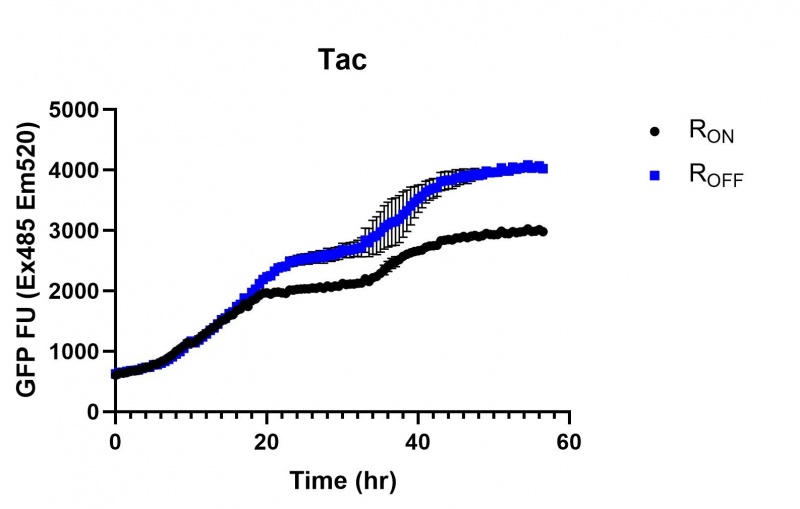
Figure 3: eGFP produced by cells transformed with the Tac riboswitch constructs [BBa_K180000] over 60 hours. We believe that the reason the Tac promoter has the opposite action of the other promoters is due to the fact that the binding of cyclic-di-GMP to the riboswitch results in shorter transcripts, allowing the RNA polymerase to produce more of the transcripts quickly and saturating the cyclic-di-GMP, resulting in a loss of inhibition of eGFP transcription.
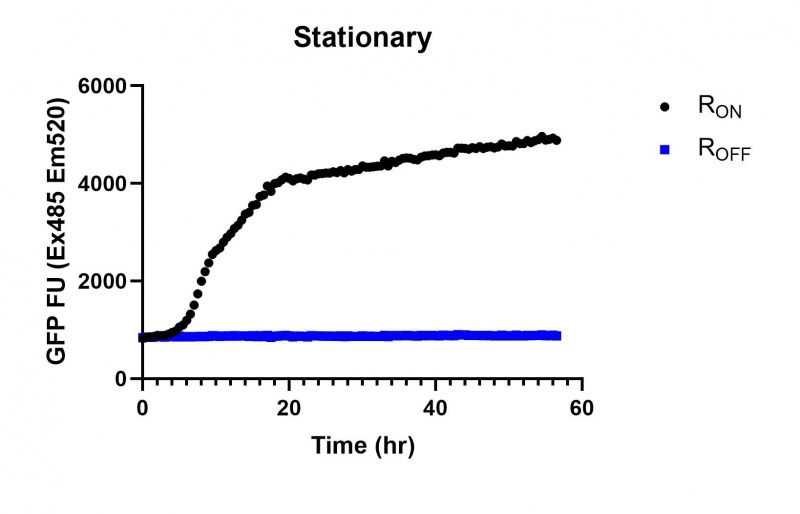
Figure 4: eGFP produced by cells transformed with the Stationary phase riboswitch constructs [BBa_K3151011] over 60 hours.
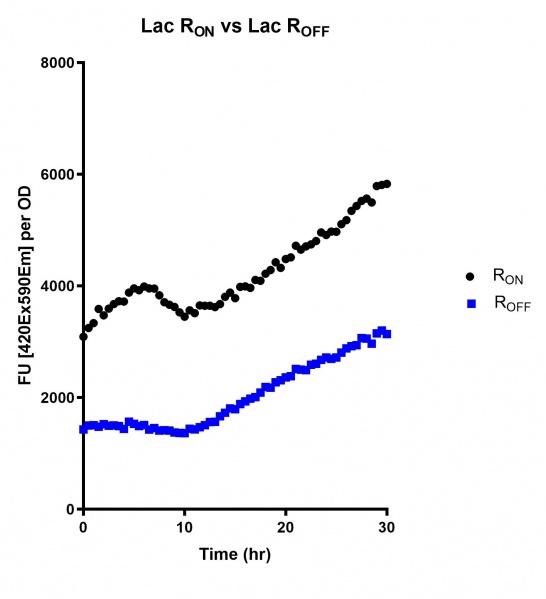
Figure 5: eGFP produced by cells transformed with the Lac riboswitch constructs [BBa_K3151028] over 30 hours. Data provided by Usyd iGEM.
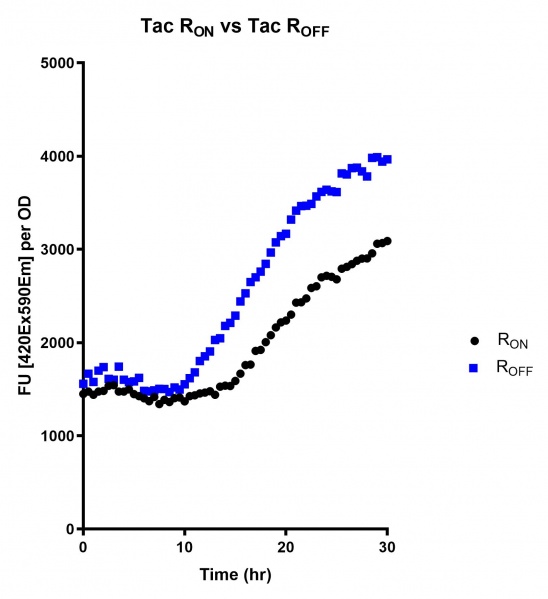
Figure 6: eGFP produced by cells transformed with the Tac riboswitch constructs [BBa_K180000] over 30 hours. Data provided by Usyd iGEM. We believe that the reason the Tac promoter has the opposite action of the other promoters is due to the fact that the binding of cyclic-di-GMP to the riboswitch results in shorter transcripts, allowing the RNA polymerase to produce more of the transcripts quickly and saturating the cyclic-di-GMP, resulting in a loss of inhibition of eGFP transcription.
In order to characterise the effect of decreased cyclic-di-GMP concentration on the riboswitch, Nissle 1917 E. coli cells transformed with the cyclic-di-GMP phosphodiesterase yhjH + RON [BBa_K3151030] and yhjH + ROFF [BBa_K3151029] constructs were spotted onto LB agar plates supplemented with Congo Red and Coomassie Blue. Biofilm formation is correlated with cyclic-di-GMP concentration[3], and can be used as a proxy for studying the effects. Induction of yhjH appeared to have no effect on the fluorescence of yhjH+RON transformants (Fig. 7A, Fig. 7C), which corroborates our hypothesis that its expression of eGFP is not influenced by the intracellular concentration of c-di-GMP. Induction of yhjH appeared to significantly increase the fluorescence of yhjH+ROFF transformants (Fig. 7B, Fig. 7D), again corroborating our hypothesis.

Figure 7: LB agar plate supplemented with Congo Red and Coomassie blue, spotted with yhjH+RON induced (A), yhjH+ROFF induced (B), yhjH+RON uninduced (C), and yhjH+ROFF uninduced (D).
In order to characterise the interaction between the function of our riboswitch and phosphodiesterases, we constructed two plasmids containing the coding sequences both the riboswitch/eGFP operon (either RON and ROFF) under the stationary phase promoter and the yhjH gene under the Lac promoter. We assembled these plasmids via standard BioBrick assembly, and confirmed assembly through restriction digestion and sequence confirmation.
E. coli DH5α cells transformed with these plasmids were used to perform fluorescence assays using the BMG Pherastar plate reader to measure eGFP (Ex485nm Em520nm) and OD (600nm) shown in Figure 8. These results confirm our hypothesis that when the yhjH phosphodiesterase is induced in the ROFF construct, it increases baseline expression of eGFP relative to the construct lacking the phosphodiesterase (Fig. 4).

Figure 8: The constitutive riboswitch [BBa_K3151011] was expressed in DH5ɑ cells. The black lines represent R ON samples, the blue lines represent R OFF samples. All R ON samples were observed to express more Green Fluorescent Protein (GFP) than corresponding R OFF samples.
References:
- Dell EJ. Bottom reading of cell-based assays: direct optic approach enhances fluorescent protein and other microplate analyses. Genetic Engineering & Biotechnology News. 2012 May 1;32(9):22-3.
- Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of rpoS-dependent genes in Escherichia coli. Journal of Bacteriology. 1993 Jan 1;175(1):259-65.
- Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, & Hengge R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes & development. 2008 Sep 1;22(17):2434-46.
| None |
