Part:BBa_K3114002
Signal peptide from OmpA (secretion tag)
Usage and Biology
The OmpA signal peptide is a 21-amino acid sequence which targets fused proteins to the Sec secretion pathway in E. coli. The fused protein is not folded until it is secreted to the periplasm. The signal peptide is cleaved after residue 21 by signal peptidase after secretion (recognition sequence: Ala-X-Ala).
Translocation of recombinant proteins to the periplasm can be an effective strategy for increasing protein yield by mitigating inclusion body formation and for simplifying purification procedures in E. coli (Sørensen and Mortensen, 2005). Many different signal peptide sequences have been characterized in literature. However, there are multiple factors that influence the ability of a particular signal peptide to target the recombinant protein of interest to the periplasm (Freudl, 2018). The optimal signal peptide candidate must therefore be determined experimentally. As such, iGEM Calgary sought to create a set of signal peptide parts so that teams may test multiple candidates in their genetic constructs to obtain optimal secretion. The parts were designed to be Golden Gate-compatible based on the MoClo standard for ease in creating the genetic constructs required to screen for the optimal candidate (Weber et al., 2011). They are also compatible with the BioBrick RFC[10] standard.
This signal peptide sequence originates from the Outer membrane protein A (OmpA) protein in E. coli (Collier, 1994). Other signal peptides within this collection include:
- DsbA signal peptide (BBa_K3114000)
- MalE signal peptide (BBa_K3114001)
- PhoA signal peptide (BBa_K3114003)
- TorA signal peptide (BBa_K3114005)
- YcbK signal peptide (BBa_K3114004)
Design
When designing this part and the rest of our collection, we were interested in creating parts that could be used in Golden Gate assembly right out of the distribution kit without the need to first domesticate them in a Golden Gate entry vector. As such, these parts are not compatible with the iGEM Type IIS RFC[1000] assembly standard because we included the BsaI restriction site and MoClo standard fusion site in the part’s sequence.
As per the MoClo standard, the 5’ signal peptide fusion sequence included in this part is AATG, and the 3’ signal peptide fusion sequence is AGGT (Weber et al., 2011).
This part includes a start codon. A Gly-Gly spacer sequence was added to the end of the signal peptide sequence in order to ensure that the fused protein of interest will be in-frame following Golden Gate assembly.
This sequence has been codon optimized for high expression in E. coli.
Characterization
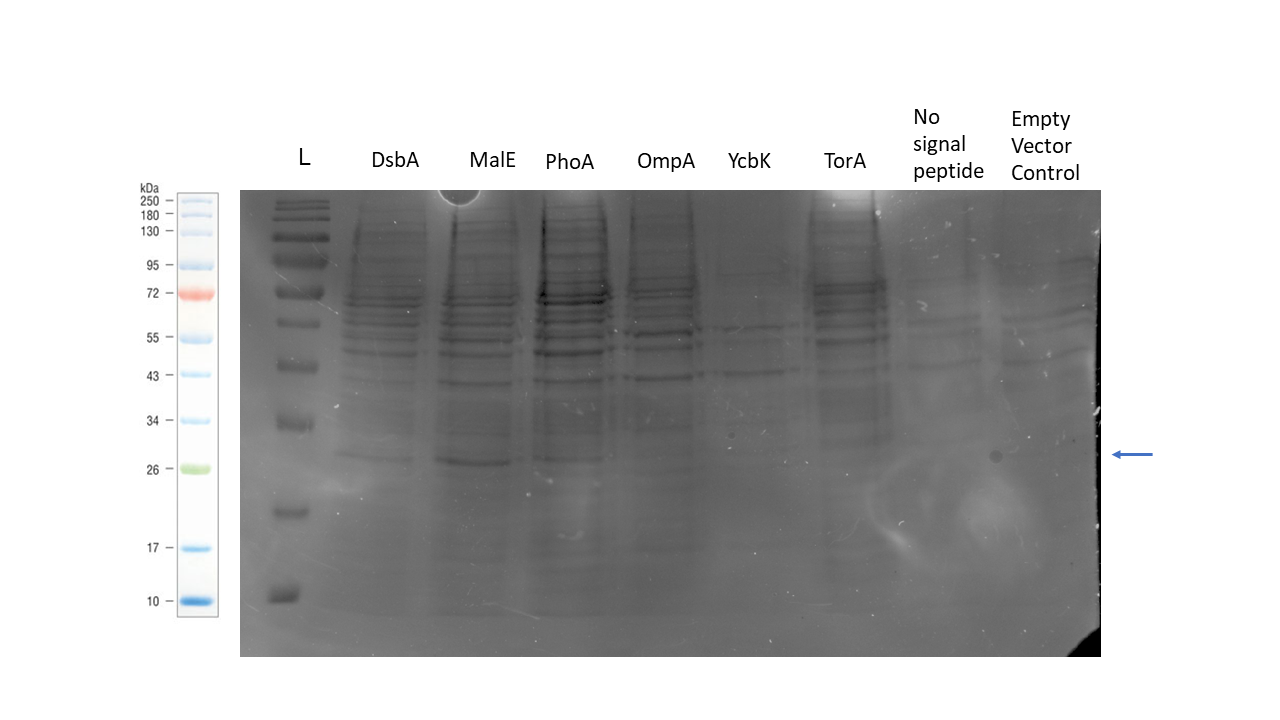
Each of the signal peptides in this collection was fused to the water-soluble chlorophyll binding protein 6GIX in a genetic circuit under the control of a T7 inducible promoter. For more information, please see iGEM Calgary's parts page.

The SDS-PAGE gel below shows the protein recovered from the periplasm for each of our genetic constructs as per our periplasmic protein isolation protocol. The construct with the MalE signal peptide showed the strongest band of the correct size, but bands are also visible for the DsbA and PhoA signal peptide samples.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1
Illegal BsaI.rc site found at 79
References
Collier, D. N. (1994). Escherichia coli signal peptides direct inefficient secretion of an outer membrane protein (OmpA) and periplasmic proteins (maltose-binding protein, ribose-binding protein, and alkaline phosphatase) in Bacillus subtilis. Journal of Bacteriology, 176(10), 3013–3020. https://doi.org/10.1128/jb.176.10.3013-3020.1994
Freudl, R. (2018, March 29). Signal peptides for recombinant protein secretion in bacterial expression systems. Microbial Cell Factories. BioMed Central Ltd. https://doi.org/10.1186/s12934-018-0901-3
Sørensen, H. P., & Mortensen, K. K. (2005). Advanced genetic strategies for recombinant protein expression in Escherichia coli. Journal of Biotechnology, 115(2), 113–128. https://doi.org/10.1016/j.jbiotec.2004.08.004
Weber, E., Engler, C., Gruetzner, R., Werner, S., & Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS ONE, 6(2). https://doi.org/10.1371/journal.pone.0016765
//proteindomain/localization
| None |