Part:BBa_K3001011
Lbu crRNA targeting GFP with T7 promoter
This CRISPR RNA (crRNA) is designed to interact with Cas13a isolated from Leptotrichia buccalis (Lbu). It is designed to target a segment of GFP mRNA transcript. It is designed to be inducibly expressed under a T7 promoter and was used for in vivo experiments in E. coli BL21(DE3). Experiments were conducted in the plasmid pUC19.
We completed a dual plasmid transformation. To confirm that we had both plasmids present we completed a digest test because PCR amplification from the colony was not working. This should separate our construct from the plasmid confirming that both parts are present. In figure 1, only linearized cuts are seen. Due to both plasmids being approximately the same size it is difficult to see. However, there are two bands present. In figure 2, only linearized cuts are seen near the 3000 bp mark. Due to both plasmids being approximately the same size it is difficult to see. However, there are two bands present. The second band at approximately 2000 bp is plasmid backbone, while the 1000 bp mark is the GFP insert. The crRNA insert (expected size is ~260 bp) is likely not seen in the gel due to having to run the gel longer to get better resolution for the upper bands.
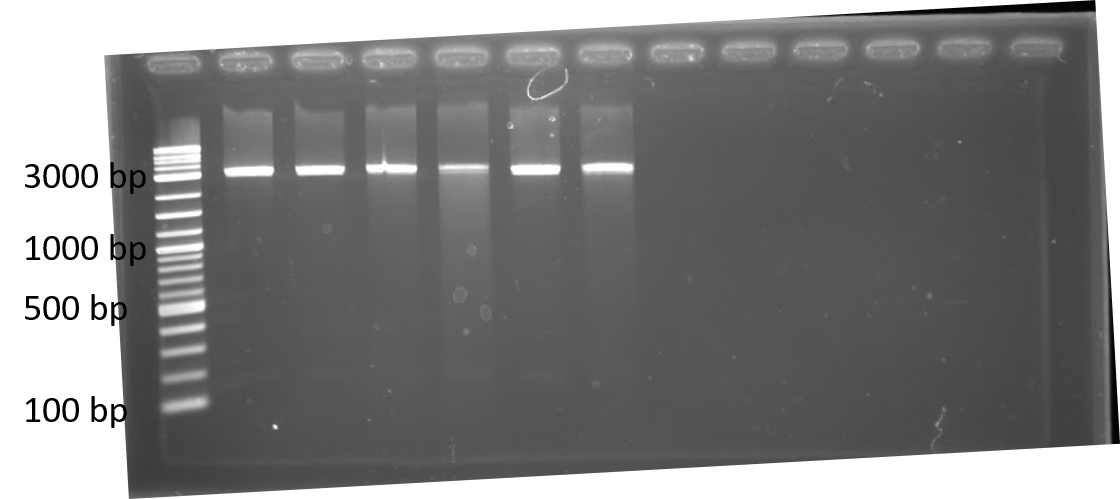 Figure 1. 1% agarose of restriction digests using PstI and EcoRI from Escherichia coli BL21(DE3) cells containing plasmids for RFP and either Lbu crRNA or Lwa crRNA. Left to right: lane 1: 1 kb ladder; lane 2: Lwa crRNA colony 1; lane 3: Lwa crRNA colony 2; lane 4: Lwa crRNA colony 3; lane 5: Lbu crRNA colony 1; lane 6: Lbu crRNA colony 2; lane 7: Lbu crRNA colony 3; lanes 8-13: empty.
Figure 1. 1% agarose of restriction digests using PstI and EcoRI from Escherichia coli BL21(DE3) cells containing plasmids for RFP and either Lbu crRNA or Lwa crRNA. Left to right: lane 1: 1 kb ladder; lane 2: Lwa crRNA colony 1; lane 3: Lwa crRNA colony 2; lane 4: Lwa crRNA colony 3; lane 5: Lbu crRNA colony 1; lane 6: Lbu crRNA colony 2; lane 7: Lbu crRNA colony 3; lanes 8-13: empty.
 Figure 2. 1% agarose of restriction digests using PstI and EcoRI from Escherichia coli BL21(DE3) cells containing plasmids for GFP and Lbu crRNA, Lwa crRNA, Lba crRNA, or Lsh crRNA. Left to right: lane 1: Lwa crRNA colony 1; lane 2: Lwa crRNA colony 2; lane 3: Lwa crRNA colony 3; lane 4: Lwa crRNA colony 4; lane 5: Lsh crRNA colony 2; lane 6: Lba crRNA colony 1; lane 7: Lba crRNA colony 2; lane 8: Lba crRNA colony 3; lane 9: Lbu crRNA colony 1; lane 10: Lbu crRNA colony 2; lane 11: Lbu crRNA colony 3; lane 12: Lbu crRNA colony 4; lane 13: 1 kb ladder.
Figure 2. 1% agarose of restriction digests using PstI and EcoRI from Escherichia coli BL21(DE3) cells containing plasmids for GFP and Lbu crRNA, Lwa crRNA, Lba crRNA, or Lsh crRNA. Left to right: lane 1: Lwa crRNA colony 1; lane 2: Lwa crRNA colony 2; lane 3: Lwa crRNA colony 3; lane 4: Lwa crRNA colony 4; lane 5: Lsh crRNA colony 2; lane 6: Lba crRNA colony 1; lane 7: Lba crRNA colony 2; lane 8: Lba crRNA colony 3; lane 9: Lbu crRNA colony 1; lane 10: Lbu crRNA colony 2; lane 11: Lbu crRNA colony 3; lane 12: Lbu crRNA colony 4; lane 13: 1 kb ladder.
Initially, our team had planned on doing a triple plasmid transformation of our target fluorescent protein (GFP), crRNA, and Cas13a to test if our system would work in vivo . Additionally, we wanted to have RFP as the fluorescent protein to serve as a specificity control instead of transforming the plasmid containing GFP. We were unsuccessful in getting all three plasmids to transform, but did succeed in getting the fluorescent proteins and crRNA containing plasmids to transform together.
As an alternative experiment, our team grew cells that expressed the Lbu and Lwa Cas13a protein overnight. We also grew cells that expressed dual plasmids; GFP and crRNA Lwa; GFP and crRNA Lbu; RFP and crRNA Lwa; and RFP and crRNA Lbu. We then lysed the cells that expressed the Cas13a proteins using a French Press and clarified the lysate via centrifugation. Following this, our team pipetted in a 1:1 ratio of clarified cell lysate: fluorescent protein and crRNA into a 96 well plate. This allowed us to observe if there would be an effect from the CRISPR Cas13a system on the fluorescent proteins. We observed that in our optical density data, both dual plasmid systems for GFP and RFP had stunted growth in comparison to only E. coli cells expressing GFP or RFP or no plasmid (Figure 3C). Adding the lysate may have caused the death of the culture. We neglected to include replicates of the dual plasmid system without adding lysate to observe how that grew. This would be beneficial for any future experiments. Alternatively, there may have been some effect of the protein in the lysate on the GFP fluorescence (Figure 3B). However, we are unsure of the specificity due to the potential of the RFP not folding correctly in vivo as demonstrated by the substantial standard deviation seen in our replicates (Figure 3A).
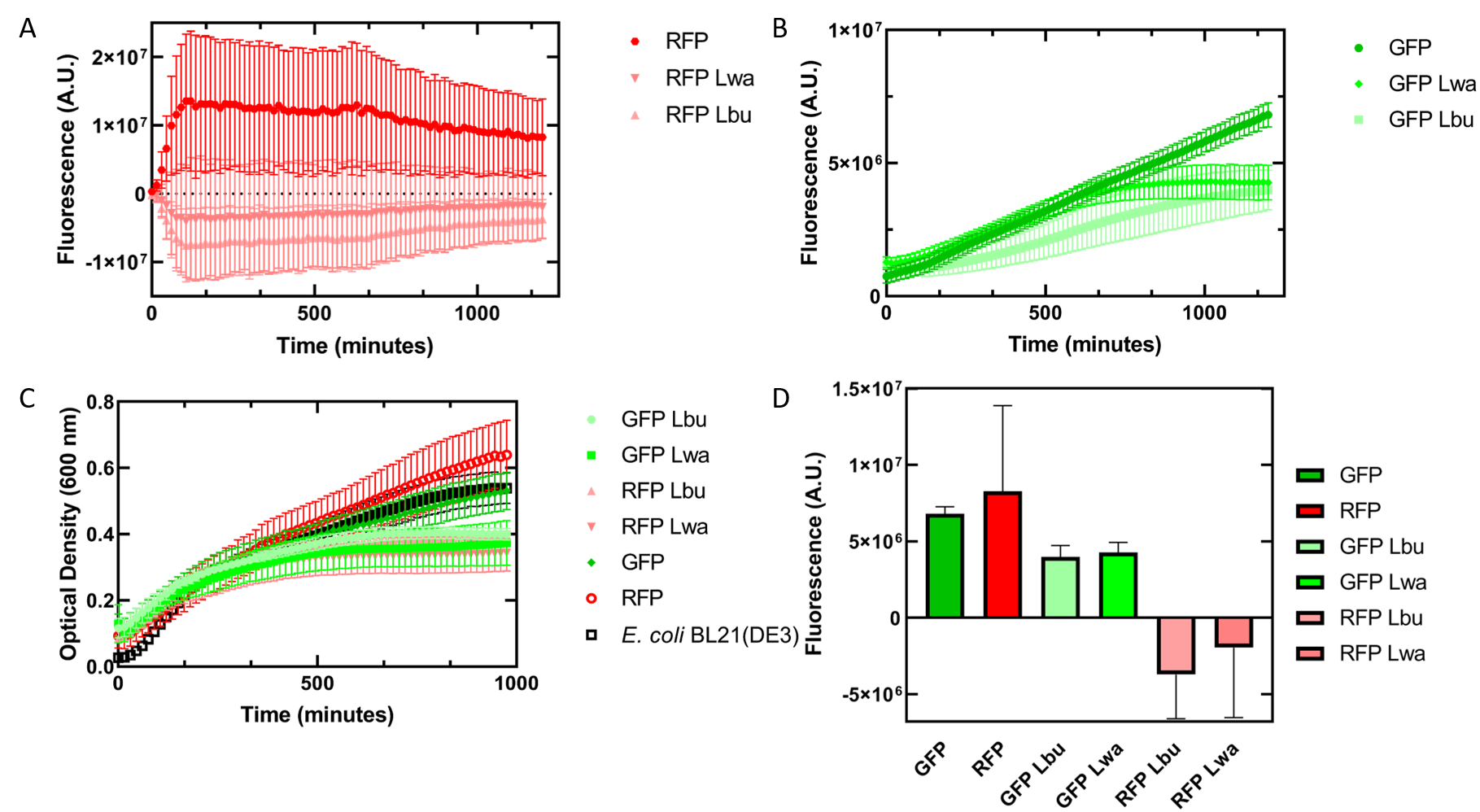 Figure 3. In vivo fluorescence assay of E. coli BL21(DE3) cells containing fluorescent protein and crRNA plasmids and E. coli Rosetta(DE3) cell lysate of overexpressed Cas13a proteins. This assay was conducted with 3 biological replicates and 3 technical replicates. (A) Fluorescence of E. coli cells containing only an RFP expressing plasmid, or dual plasmid expression of RFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. RFP excitation was at 558 nm and emission at 583 nm. (B) Fluorescence of E. coli cells containing only a GFP expressing plasmid, or dual plasmid expression of GFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. GFP excitation was at 475 nm and emission at 508 nm. (C) Optical density of E. coli cells expressing GFP, RFP, dual plasmid systems mentioned previously, or only E. coli BL21(DE3) cells with absorbance measured at 600 nm. (D) Relative fluorescence at maximum excitation at 81 minutes. GFP excitation was at 475 nm and emission at 508 nm and RFP excitation was at 558 nm and emission at 583 nm.
Figure 3. In vivo fluorescence assay of E. coli BL21(DE3) cells containing fluorescent protein and crRNA plasmids and E. coli Rosetta(DE3) cell lysate of overexpressed Cas13a proteins. This assay was conducted with 3 biological replicates and 3 technical replicates. (A) Fluorescence of E. coli cells containing only an RFP expressing plasmid, or dual plasmid expression of RFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. RFP excitation was at 558 nm and emission at 583 nm. (B) Fluorescence of E. coli cells containing only a GFP expressing plasmid, or dual plasmid expression of GFP and crRNAs from Lwa and Lbu that target GFP and the respective cell lysate containing the appropriate Cas13a. GFP excitation was at 475 nm and emission at 508 nm. (C) Optical density of E. coli cells expressing GFP, RFP, dual plasmid systems mentioned previously, or only E. coli BL21(DE3) cells with absorbance measured at 600 nm. (D) Relative fluorescence at maximum excitation at 81 minutes. GFP excitation was at 475 nm and emission at 508 nm and RFP excitation was at 558 nm and emission at 583 nm.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
