Difference between revisions of "Chassis/B.subtilis key parts"
m (→AmyE Integration Biobricks) |
m (→Constitutive Promoters) |
||
| Line 23: | Line 23: | ||
[[Image:TestingPromoters.PNG|600px|center]] | [[Image:TestingPromoters.PNG|600px|center]] | ||
| − | The pVeg promoter and spoVG ribosome binding site (RBS) Biobrick ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K143053 BBa_K143053]) has been characterised for expression of both GFPmut3b ([https://parts.igem.org/wiki/index.php?title=Part:BBa_E0040 BBa_E0040]) and mRFP1 ([https://parts.igem.org/wiki/index.php?title=Part:BBa_E1010 BBa_E1010]). This data not only shows the parts are both working but once further parts have been characterized provides a basis for comparison. Further promoter and RBS combinations are currently being characterized, in addition to the construction of calibration curves to normalise the units of fluorescence to molecules of fluorescent proteins. | + | The pVeg promoter and spoVG ribosome binding site (RBS) Biobrick ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K143053 BBa_K143053]) has been characterised for expression of both GFPmut3b ([https://parts.igem.org/wiki/index.php?title=Part:BBa_E0040 BBa_E0040]) and mRFP1 ([https://parts.igem.org/wiki/index.php?title=Part:BBa_E1010 BBa_E1010]). This data not only shows the parts are both working but once further parts have been characterized provides a basis for comparison. Further promoter and RBS combinations are currently being characterized, in addition to the construction of calibration curves to normalise the units of fluorescence to molecules of fluorescent proteins. For more information about this biobrick please see [http://2008.igem.org/Team:Imperial_College Imperial College 2008 iGEM project.] |
==Multi-host vector pTG262== | ==Multi-host vector pTG262== | ||
Revision as of 02:00, 30 October 2008
| B.subtilis strains | B. subtilis chassis characterisation | B. subtilis specific parts | Key Parts and Devices |
Bacillus subtilis Key Parts
This page is aimed to highlight some of the key biobricks for B. subtilis that are available on the Registry of Standard Biological Parts.
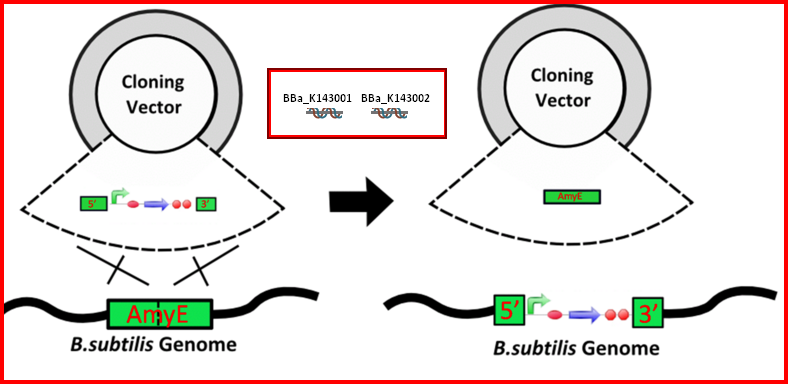
AmyE Integration Biobricks
The AmyE integration bricks (BBa_K143001 and BBa_K143002) are composed of two biobricks that can be added to the 5' and 3' ends of a construct to allow integration into the B. subtilis genome. These biobricks have been successfully used within the constructs BBa_K143079 and BBa_K143082for integration. The possible advantages of integration are within stability and control of copy number. For more information about this biobrick please see [http://2008.igem.org/Team:Imperial_College Imperial College 2008 iGEM project.]
Constitutive Promoters
The pVeg promoter and spoVG ribosome binding site (RBS) Biobrick (BBa_K143053) has been characterised for expression of both GFPmut3b (BBa_E0040) and mRFP1 (BBa_E1010). This data not only shows the parts are both working but once further parts have been characterized provides a basis for comparison. Further promoter and RBS combinations are currently being characterized, in addition to the construction of calibration curves to normalise the units of fluorescence to molecules of fluorescent proteins. For more information about this biobrick please see [http://2008.igem.org/Team:Imperial_College Imperial College 2008 iGEM project.]
Multi-host vector pTG262
This plasmid was derived from pTG262 by insertion of a biobrick encoding Red Fluorescent Protein between the unique EcoRI and PstI restriction sites. This eliminated the only native XbaI site present in the plasmid, and since there are no native SpeI sites, the resulting plasmid is a compliant BioBrick vector. New biobricks can be inserted into this vector by replacement of the RFP biobrick, and selection of the white colonies.The parent plasmid pTG262 has a multi host replication origin and replicates in a range of gram positive and gram negative bacteria including E. coli, Bacillus subtilis, and Lactobacillus and Lactococcus spp. We hope that it will also replicate in other Gram negative bacteria. At the time of writing we have successfully transformed E. coli and B. subtilis with this construct, selecting with chloramphenicol (15 and 10 mg/l respectively)