Part:BBa_K361000
Rci site-specific recombinase
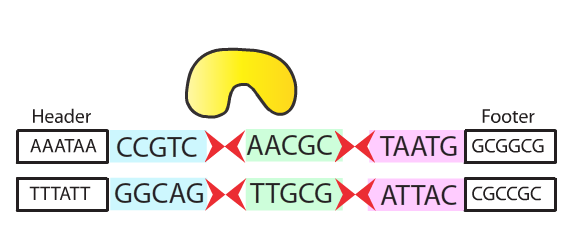
This part is responsible for the functioning protein - Rci site-specific recombinase (384-amino acid) in Shufflon system, a site specific DNA recombination would occur between two specific repeat sites.
With the expression of this gene, the DNA sequence would be shuffled randomly according to the specific repeat sites recognized by the recombinase.
Recombinase activity| Intramolecular inversion frequency | 0.062 |
|---|---|
| Intramolecular deletion frequency | 0 |
| Intramolecular recombination | 0 |
The inversion frequency is affected by the length of flanking DNA sequence in vivo. Please refer to BBa_K361001
Usage and Biology
Usage:
- The Rci site-specific recombinase is utilized by the bacteria in order to adapt the environmental changes by rearranging their gene pattern, the DNA shuffling effect may provide selective advantages in the new environment to survive.
- Integrated into our system, the Rci site-specific recombinase now functions as an encryption system - shuffling information that are stored in DNA.
Biology:
- In vitro study of conditions affecting recombination:
1. Additions of magnesium ion, EDTA, or ATP had little effect on recombination
→Neither an energy source nor divalent cations are necessary.
2. The inversion was sensitive to KCl concentration
→Maximal inversion activity at a KCl concentration of 80 mM.
3. The inversion reaction showed a broad pH optimum between 7.6 and 8.5.
4. The inversion reaction has a higher reaction activity at 42 oC as compared with that of 25 oC and 30 oC.
5. Negatively supercoiled DNA substrate was required for Rci protein. Little to zero recombination was observed in relaxed or linear DNA.
- In vitro study of recombination:
About 15-20 % inversion / recombination
- Reference:
[1] Gyohda, A. & Komano, T. (2000). Purification and Characterization of the R64 Shufflon-Specific Recombinase. Journal of Bacteriology, 182 (10), 2787-2792.
[2] Gyohda, A., Zhu, S., Furuya, N. & Komano, T. (2005). Asymmetry of Shufflon-specific Recombination Sites in Plasmid R65 Inhibits Recombination between Direct sfx Sequences. The Journal of Biological Chemistry, 281 (30), 20772-20779.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 362
Illegal NgoMIV site found at 857 - 1000COMPATIBLE WITH RFC[1000]
//function/recombination
| biology | active wide type enzyme |
| function | Site-specific recombination |
| protein |