Part:BBa_I13453
Pbad promoter
PBad promoter from I0500 without AraC.
Usage and Biology
Has been used as a second promoter in a system containing BBa_I0500 (PBad+AraC). In this system, it showed behavior qualitatively indistinguishable from the BBa_I0500 copy of PBad. Has not been tested independent of AraC. A second part, BBa_I13458, should allow decoupling of PBad and AraC.
See this OpenWetWare article on [http://openwetware.org/wiki/Titratable_control_of_pBAD_and_lac_promoters_in_individual_E._coli_cells#pBAD_promotersOpenWetWare pBAD and lac promoters] for additional usage and biology information
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 125
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 65
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
For the 2009 iGEM competition, British_Columbia characterized BBa_I13453 in the context of a pBAD promoter family. For the results of this characterization, see here.
For the 2010 iGEM competition, Tec-Monterrey characterized BBa_I13453 again, using a construct of GFP reporter after the pBad promoter. The transfer function was modeled with a Hill equation.
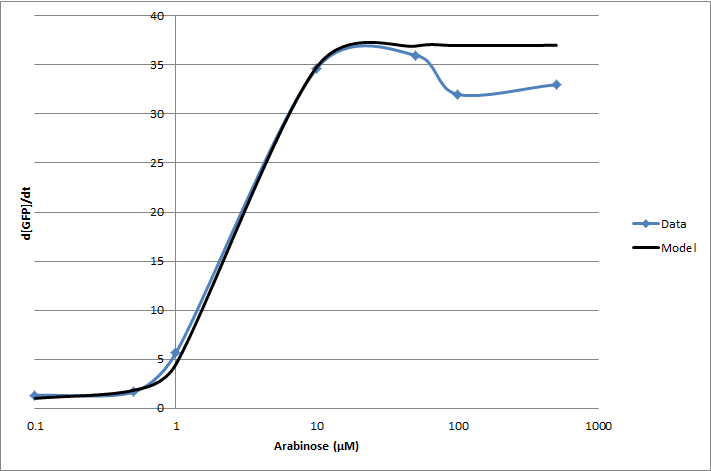
(d[GFP]/dt)/OD600 = C+A*Xn/(Xn+Kn)
| Experiment | Characteristic | Value |
|---|---|---|
| Transfer Function | Basal rate (C) | 1 d[GFP]/dt |
| Gain (A) | 36 d[GFP]/dt | |
| Hill coefficient (n) | 2.16 | |
| Switch Point (K) | 2.8 [ara] (µM) |
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/positive
//classic/regulatory/uncategorized
| negative_regulators | |
| positive_regulators | 1 |

 1 Registry Star
1 Registry Star