Part:BBa_I721001
Lead Promoter
Lead Promoter (Part 4) messed up EcoRI site
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 80
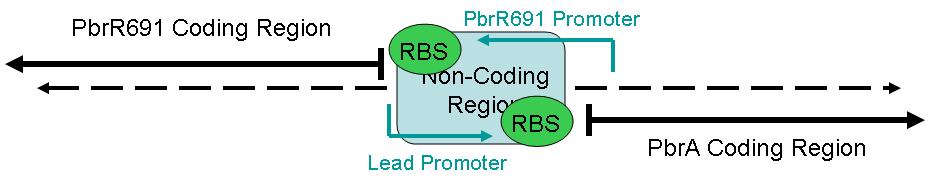
Part BBa_1721001 is the Lead Promoter.
Description
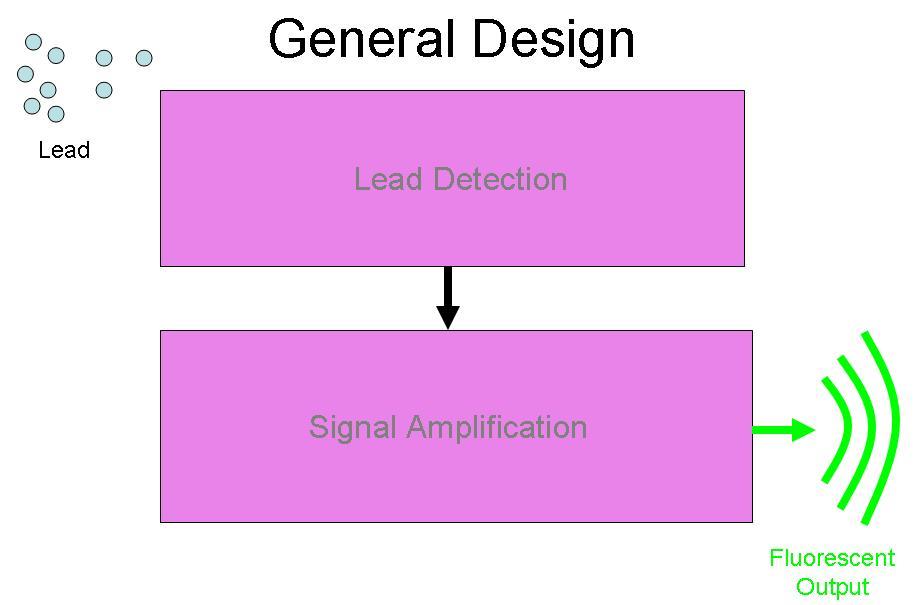
This part is designed to enable Lead Detection in a cell. When Lead (Pb +2) enters the cell, it will couple with the Lead Binding protein (NOT IN THIS PART), forming a dimer. This dimer will then bind to the Lead Promoter, which is contained within this part, BBa_I721001. BBa_I721001 contains the Lead Promoter sequence, and and must be combined with a functional Lead Binding Protein part for it to work.
This sequence was obtained using PCR from the genome of Ralstonia metallidurans. Biobrick cut sites were added using specific primers during PCR.
This part is not known to function. "PbrR691/promoter combinations in presence of lead nitrate give no GFP production and no AHL production compared to control. "
The figure below illustrates the layout of this sequence.
This part contains one section: A sequence for a Lead Promoter.
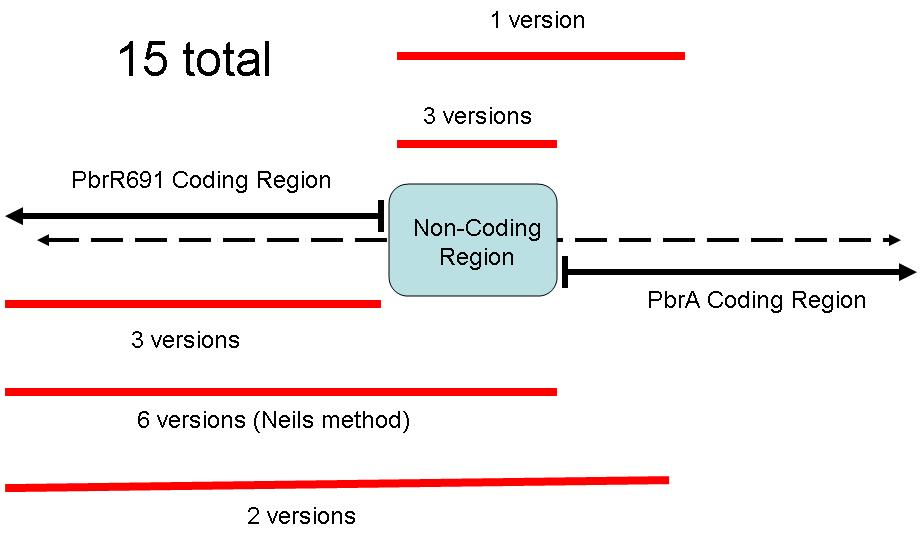
Because of the different coding regions contained in this sequence, the Brown iGEM Team chose 15 different regions of the Pbr691 and PbrR691 sequences to extract. We attempted to create Biobricks from each of these 15 different sequences. Some contained only the PbrR Lead Binding Protein, some contained only the Pbr Promoter, and some parts contained both. The following diagram illustrates these different parts.
BBa_I721001 contains the Lead Promoter region.
BBa_I721001 was designed to be a part of the Brown University Cellular Lead detector device. The BBa_I721001 Lead Promoter is intended to be combined with a Lead Binding Protein Part. Together, this Lead Detection System was designed to be attached to the Amplification system by Imperial College, (BBa_J37015), with GFP as the output of the device. The schematic for this project is below:
Contribution
- Group: iGEM24_HongKong-JSS
Lead biosensor
Our team used this part to construct a lead biosensor with E. coli. We aimed to create biosensors visible to the naked eye without special equipment, so we employed the dTomato chromoprotein, previously optimized by our team, as the reporter gene (BBa_K5152004).
We successfully validated our biosensor's ability to detect lead (100 µM lead (II) nitrate). The medium turns red due to dTomato expression. We obtained pellets from 1 mL of biosensor cultures after spinning at 8,000 g for 2 minutes.


The colour signal in the pellet becomes distinguishable after 12 hours of incubation, while the culture's signal is significant after 18 hours with lead. However, leaky red expression is observed if incubation exceeds 18 hours.
We also demonstrated that the reporter is concentration-dependent; the color intensity increases with higher lead concentrations. The figure below shows our biosensor after incubating with 0, 0.01, 0.1, 1, and 10 µM lead (II) nitrate solutions for 24 hours. The color differences are easily distinguishable.

This suggests that our biosensor design is effective and could have broad applications.
//direction/forward
//function/sensor/metal
//promoter
//regulation/positive
//rnap/prokaryote/ecoli/sigma70
| negative_regulators | |
| positive_regulators | 1 |

 1 Registry Star
1 Registry Star