Part:BBa_K3113009
L7Ae
L7Ae is a 50S ribosomal protein found in Archea. It is a multifunctional RNA-binding protein that recognizes the K-turn motif in ribosomal RNA, the RNA component of RNase P, box H/ACA, box C/D, box C'/D' sRNAs. This RNA-binding protein can be used as an adapter for RNA binding in the extracellular vesicles.
Usage
The informational readout of ALiVE is RNA. To load a specific RNA into the vesicles we fused it to RNA-motifs which are bound specifically by RNA binding proteins. Two combinations of binding protein and motif were tested. One was the archaeal RNA binding protein L7Ae which binds to the k-turns in the box C/D snoRNP.
Biology
The archaeal L7Ae and eukaryotic 15.5kD protein homologs are members of the L7Ae/15.5kD protein family that characteristically recognize K-turn motifs found in both archaeal and eukaryotic RNAs. In Archaea, the L7Ae protein uniquely binds the K-loop motif found in box C/D.[1] The k-turn is a ubiquitous structural motif in RNA forming a very tight kink in the axis of helical RNA that plays an important role in many aspects of RNA function. L7Ae is a member of a superfamily of proteins that bind k-turns in RNA, stabilizing the tightly kinked conformation. They are extremely widespread and are important in the assembly of RNA–protein complexes central to translation, splicing and site-specific RNA modification. [2]
Literature Characterization by AFCM-Egypt
The study made Western blot analysis and confocal immunofluorescent analyses of different cell lines using the L7a (E109) antibody.

By using Ribosomal Protein L7a (E109) Antibody, the study made Western blot analyses of extracts from various cell lines.

The study used the ribosomal protein L7a (E109) antibody, which appeared green, to make a confocal immunofluorescent analysis of COS cells. They labeled actin filaments with Alexa Fluor phalloidin 555, which appear red. DRAQ5 #4084 (fluorescent DNA dye) appeared as a blue pseudocolor. The antibody stains ribosomes at their biogenesis location in nucleoli.
Characterization By Mutational Landscape by AFCM-Egypt
In order to optimize the function of our parts, we've used the concept of Directed Evolution through applying different mutations and measuring the effects of these mutations on their evolutionary epistatic fitness. As displayed in the chart below, the mutation (N80K) shows the highest epistatic fitness, while the lowest score was associated with the mutation (N99F,P91C,C92I).

Figure . An illustration of the effects of different mutations on the Epistatic Fitness of L7ae.
Experimental Characterization by AFCM-Egypt
In order to amplify this DNA part, we used PCR amplification to reach the desired concentration to complete our experiments using specific forward and reverse primers, running the parts on gel electrophoresis as this part presents in lane (P4) including CD63 an L7Ae, and then measuring the specific concentration of the running part using Real-Time PCR as shown in the following figure.

We performed the double digestion method for this part in the prefix and suffix with its specific restriction enzyme and applied this part to gel electrophoresis as shown in the following figure lane (P4).

Characterization
Secretion efficiency
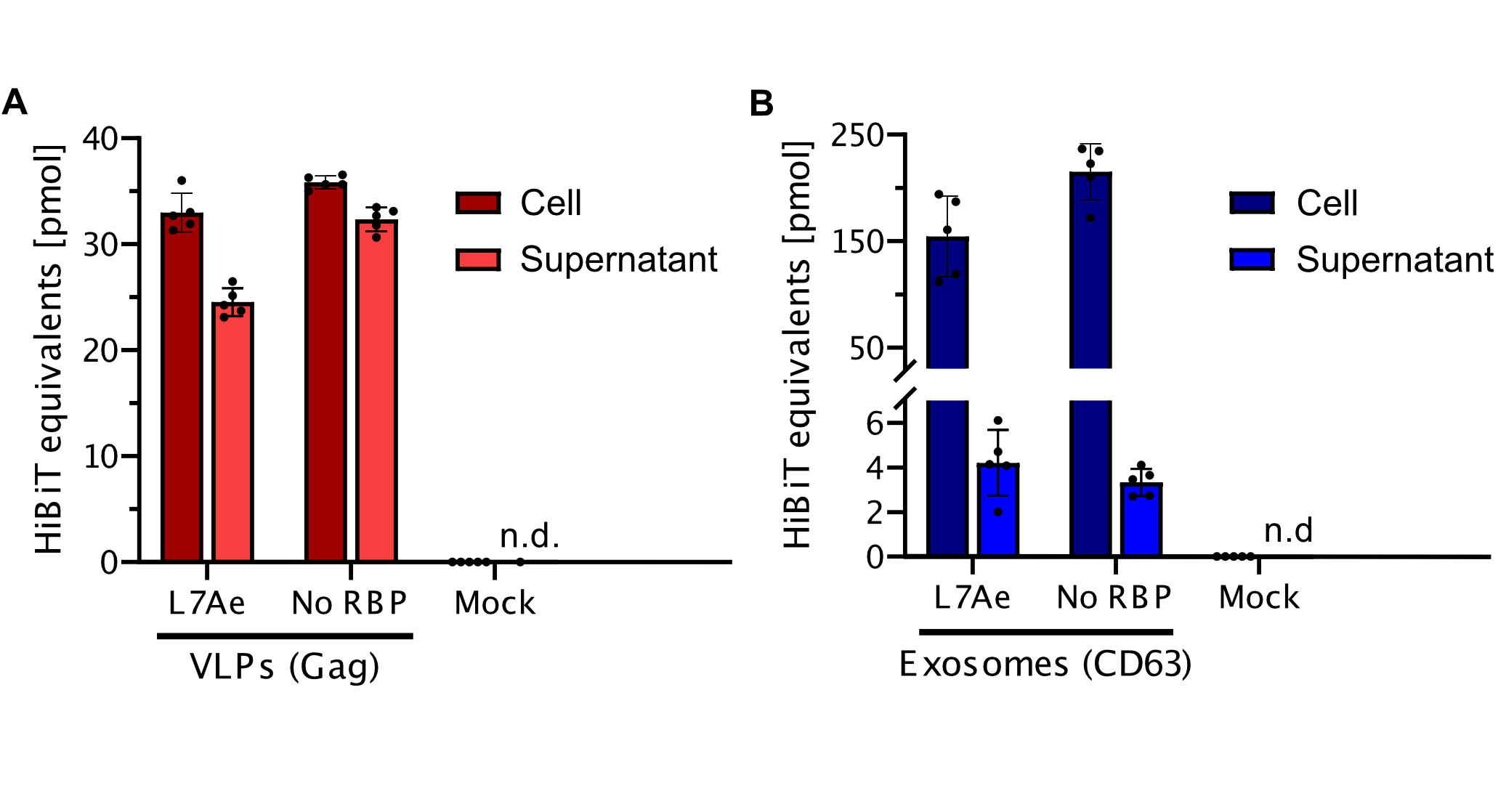
qPCR
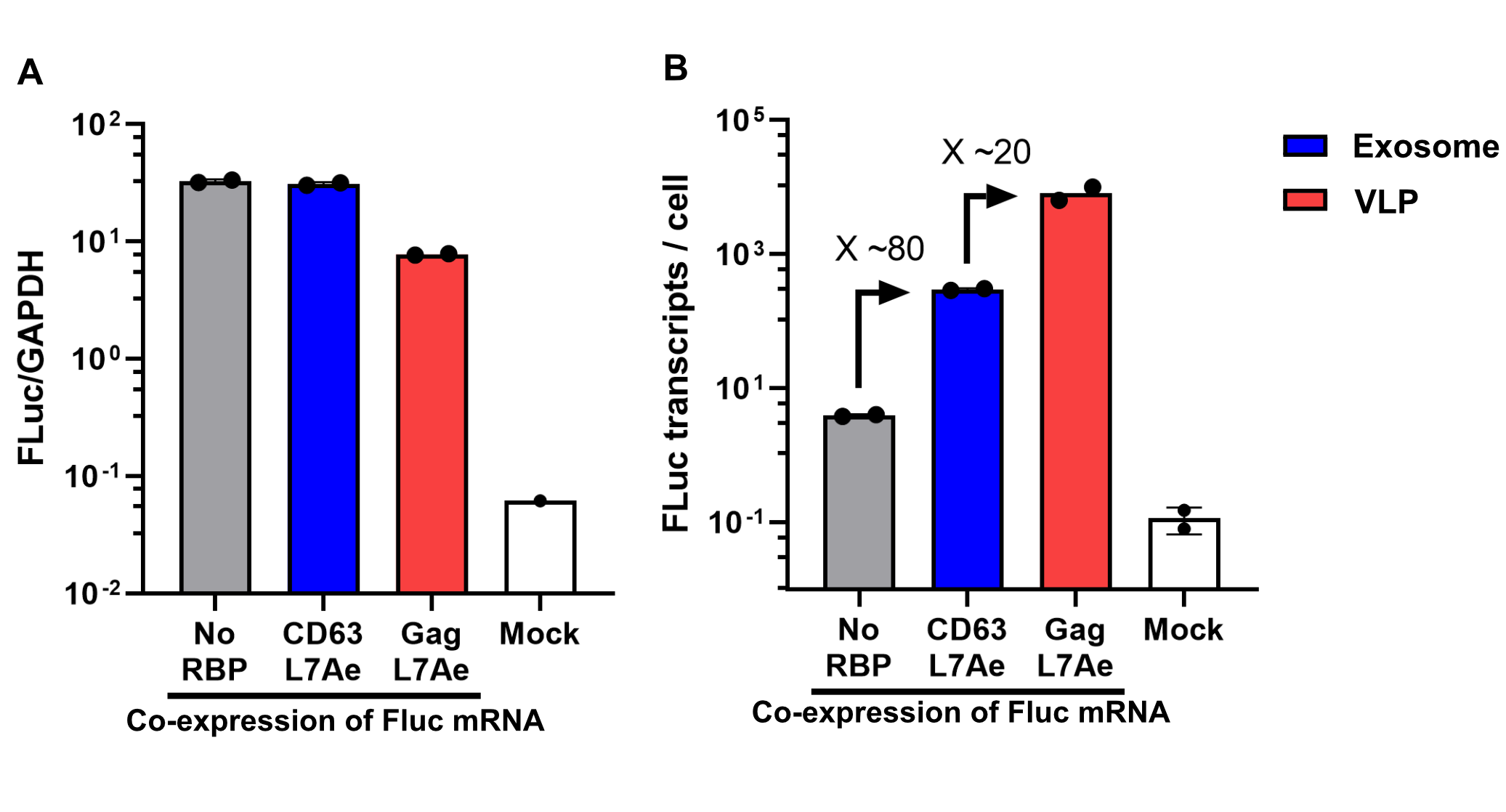
Using a coiled-coil system allowed us to successfully load the two different RNA binding proteins (RBPs) L7Ae or MCP into exosomes to export FLuc mRNA from HEK293T cells. We used the published parallel heterodimer pair P9SN:P10SN (Ljubetič et al. 2017), where P9SN was fused to the C-terminus of the exosome marker protein CD63 and P10SN N-terminal to the RNA binding proteins L7Ae or MCP. The export efficiencies of FLuc mRNA cargo measured by qPCR proved that coiled-coil mediated targeting of the RBPs L7Ae and MCP into exosomes worked. Exosomes formed from CD63-P9SN and loaded with P10SN-L7Ae or -MCP could export about 800 and 400 transcripts per cell, respectively. This also shows that the two RBPs have different export capabilities, with L7Ae exporting twice as many transcripts per cell compared to MCP.

- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Gagnon, Keith T et al. “Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-loop RNA motif.” RNA (New York, N.Y.) vol. 16,1 (2010): 79-90. doi:10.1261/rna.1692310
- ↑ Lilley, D. M. J. (2019). "The L7Ae proteins mediate a widespread and highly functional protein–RNA interaction." The Biochemist 41(2): 40-44.
| None |
