Part:BBa_K4926006
GbcAsg1
cas12a needs to bind to crRNA to be active, and the sequence of the crRNA complementary to its target DNA determines the recognition targeting of cas12a. This is the primer used for in vitro transcription to get the corresponding crRNA of the target sequence.
Usage and Biology
This part was designed for Pseudomonas aeruginosa's characteristic sequence, GbcA, to be used as a primer for in vitro transcription to obtain sgRNA.
Pseudomonas aeruginosa is a Gram-negative bacterium first isolated from wound pus in 1882[1]. Pseudomonas aeruginosa is an aerobic, long rod-shaped bacterium with only unidirectional motility. It is an opportunistically infecting bacterium and is also opportunistically infecting plants, and is so named because of the green color of the disease material such as pus and exudate after infection[2]. It is widely distributed in nature and normal human skin, intestinal tract, and respiratory tract and is one of the more common conditionally pathogenic bacteria in clinical practice[3].
In this project, we designed and developed a rapid detection method for Pseudomonas aeruginosa based on CRISPR/Cas12a technology. The principle of this detection system is to use Cas12a protein to cut specific nucleic acid sequences of bacteria as a way to detect the presence of bacteria. Since cas12a needs to bind to crRNA to be active, and the sequence of the crRNA complementary to its target DNA determines the recognition targeting of cas12a[4]. Therefore, we need to design primers to obtain the corresponding gRNA by in vitro transcription.
Cultivation, Purification, and SDS-PAGE
PCR amplification and plasmid construction
GbcA was amplified from the genome of Pseudomonas aeruginosa respectively. In order to construct our plasmids, we let the company synthesize the DNA fragment GbcA. While the pET-28a(+)-LbCas12a-6His was obtained from the company laboratories.

Figure 1: pUC57-GbcA in DH5α.
We transformed the pUC57-GbcA into the E. coli DH5α. As shown in Figure 1, we obtained the E. coli DH5α with target fragments of GbcA. To verify whether these two plasmids presented in the E. coli DH5α, we performed TAE agarose gel electrophoresis. As shown in Figure 2, we got lanes which length were the same with GbcA (646 bp). So we confirmed that the right recombinant plasmid was in the E. coli DH5α. (Figure 2).

Figure 2: Bacteria PCR of GbcA.
In vitro transcription of sgRNA
The corresponding genes of sgRNA of GbcA were obtained by PCR (isothermal amplification) as DNA templates. We verified that the sgDNA is the one we planned to target by checking the bp length (Figure 3).
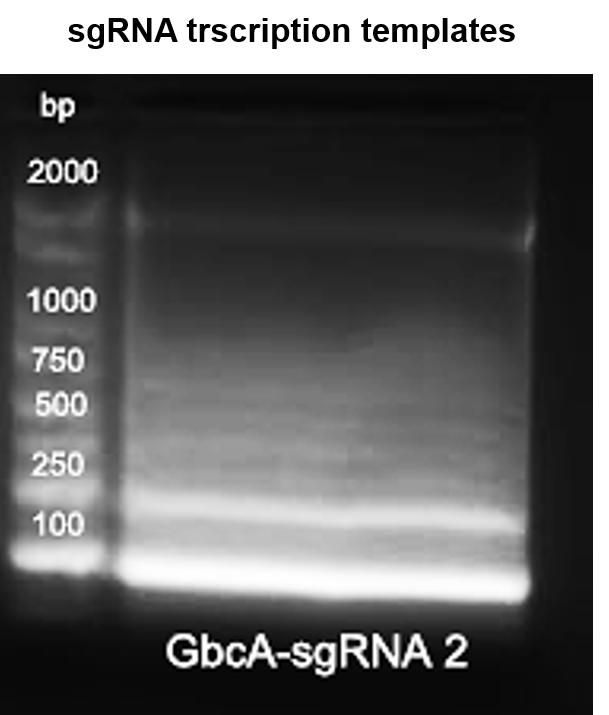
Figure 3: DNA fragment of GbcA in target sgRNA.
We then used T7 in vitro transcription kit to obtain the sgRNA of GbcA. We also checked whether the sgRNA underwent a successful transcription by taking its DNA template as a negative control. As shown in Figure 4, since sgRNA of GbcA is under 100bp, we confirmed that the T7 in vitro transcription was successful.

Figure 4: GbcA sgRNA and DNA template.
Cas-12a Protein Expression and Purification
We transformed the pET-28a(+)-LbCas12a-6His expression plasmid into E. coli BL21(DE3) and cultured overnight. Then we transferred the medium into 300mL fresh LB culture medium. When the OD600 was around 0.6, we added IPTG to induce LbCas12a expression at 16℃ for 12 hours. Then LbCas12a was extracted and purified by His-tag.

Figure 5: SDS-PAGE analysis of LbCas12a protein.
Measurement
Function Validation

Figure 6: Graph of efficiency comparison of sgRNAs.
After the testing of the fluorescence of the sgRNA after incubation, we drew this graph to show the efficiency of the different sgRNAs. In the graph, a faster decreasing slope means higher efficiency in uncoiling the plasmid. Since all the sgRNA shows a decreasing slope, all of them successfully react with the plasmids, which means our experiment succeeded. The gbc-sg1RNA decreased fastest with the highest change in slope among the same types, which means they have the highest efficiency.
Besides, we tested the reaction of the sgRNA in different concentration of bacteria liquid containing plasmid pUC57-GbcA. By this, we can find the minimum detectable concentration, and get the sensitivity of our test kit.

Figure 7: Relative fluorescence intensity of different sgRNA for GbcA (represent the sensitivity of different sgRNA).
At the point that relative fluorescence intensity gets 1000000, human eyes could identify the change in the light intensity. Therefore, the line of relative fluorescence intensity of different sgRNA that gets the 1000000 in the least time would represent the most effective sgRNA. And by analyzing the 1ul and 2ul, we can find the sensitivity of each sgRNA. After analyzing, we find out that the sgRNA2 in GbcA group is the most sensitive sgRNA.
In conclusion, each sgRNA has their own advantages and disadvantages. Therefore, we would not focus on any single type of sgRNA to make our product. We will try to mix these sgRNA together in a specific concentration ratio to get the highest efficiency and sensitivity in detecting the pathogen.
Reference
- Hassoun-Kheir N, Guedes M, Ngo Nsoga MT, Argante L, Arieti F, Gladstone BP, Kingston R, Naylor NR, Pezzani MD, Pouwels KB, Robotham JV, Rodríguez-Baño J, Tacconelli E, Vella V, Harbarth S, de Kraker MEA; PrIMAVeRa Workpackage 1. A systematic review on the excess health risk of antibiotic-resistant bloodstream infections for six key pathogens in Europe. Clin Microbiol Infect. 2023 Oct 4:S1198-743X(23)00419-6. doi: 10.1016/j.cmi.2023.09.001. Epub ahead of print. PMID: 37802750.
- Dehari D, Kumar DN, Chaudhuri A, Kumar A, Kumar R, Kumar D, Singh S, Nath G, Agrawal AK. Bacteriophage entrapped chitosan microgel for biofilm-mediated polybacterial infection in burn wounds. Int J Biol Macromol. 2023 Oct 4:127247. doi: 10.1016/j.ijbiomac.2023.127247. Epub ahead of print. PMID: 37802451.
- Zhao Z, Luo YH, Wang TH, Sinha S, Ling L, Rittmann B, Alvarez P, Perreault F, Westerhoff P. Phenotypic and Transcriptional Responses of Pseudomonas aeruginosa Biofilms to UV-C Irradiation via Side-Emitting Optical Fibers: Implications for Biofouling Control. Environ Sci Technol. 2023 Oct 6. doi: 10.1021/acs.est.3c04658. Epub ahead of print. PMID: 37802050.
- Fang, L., Yang, L., Han, M., Xu, H., Ding, W., & Dong, X. (2023). CRISPR-cas technology: A key approach for SARS-CoV-2 detection. Frontiers in bioengineering and biotechnology, 11, 1158672. https://doi.org/10.3389/fbioe.2023.1158672
