Part:BBa_K3868102
pET-Alloferon-1-eGFP
pET--Alloferon-1-eGFP plasmid contains pT7, lacO,Alloferon-1, thrombin, eGFP-His-tag etc.Based on the plasmid of pET-24a, the C-terminal of Alloferon-1 was fused with eGFP in order to characterize the yield of Alloferon-1. Moreover, in order to purify the Alloferon-1, the enzyme loci of thrombin was inserted between Alloferon-1 and eGFP, and the label of 6*His was inserted at the end of eGFP, yielding the plasmid of pET-Alloferon-1-eGFP. The fluorescence intensity of eGFP could be used to indicate the expression level of Alloferon-1.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Based on the plasmid of pET-24a. The C-terminal of Antibacterial peptides(AMP) was fused with eGFP in order to characterize the yield of AMP. The pET-24a was provided by our PI. Moreover, in order to purify the AMP, the enzyme loci of thrombin was inserted between AMP and eGFP, and the label of 6*His was inserted at the end of eGFP, yielding the plasmid of pET-AMP-eGFP. As a result, the pET-AMP-eGFP plasmids could not only indicate the expression level of AMPs by the fluorescence intensity of eGFP, but also be good for later purification and separation with His-Tag and the enzyme loci of thrombin. (Fig. 1)

- Fig 1. The Schematic diagram of pET-AMP-eGFP
Improve From NJXDF-CHN 2022
Group: iGEM22_NJXDF-CHN
Author: Yang Gu
Overview
We reviewed the relevant work of NNU-China in 2021. This project utilized the cytosine base editor (CBE) and CRISPR/Cas9 technology to construct a ribosome binding site (RBS) library of T7 RNA polymerase (RNAP), which was applied for rapid screening of expression hosts suitable for antimicrobial peptide (AMP) production. However, we found that not all of the AMPs involved in the optimization had significant increases in yield. We hypothesized that the RBS sequence of the target gene in the pET plasmid also plays a decisive role in the recombinant protein yield. Therefore, we selected the relevant components used by the team, CBE (BBa_K3868097) and Alloferon-1 (BBa_K3868102), for enhancement. This enhancement is expected to be extended to the optimization of various pET series plasmids.
Design
To prevent over-editing, we replace the promoter in BBa_K3868097 with a temperature-sensitive promoter (obtained by BBa_Q04510), which can be used to manipulate the start and end of editing by temperature switching (PL-CBE) (Fig 1). Notably, the subsequent recombinant plasmids were constructed on the basis of the original plasmid pSC101 (CmR) or pET-24a (KanR) using the restriction-ligation method. Further, the recombinant plasmids were all transformed by electro-transformation. In brief, Escherichia coli BL21 (DE3) or derived strains were cultured in 4 mL LB medium (OD600 reached at 0.6-0.8). The above bacterial liquid was centrifuged at 4,000 rpm for 10 minutes at 4°C. Next, add 1 mL glycerol (10%), pipet up and down slightly to mix thoroughly. And centrifuge at 4,000rpm for 10 minute at 4℃ temperature. Decant or aspirate medium and discard. The above steps need to be repeated once. Add 100 μL glycerol (10%) and recombinant plasmid or linearized fragment (100 ng or more) to the bacteria, and transfer to the electro-rotor cup. After electroshock (1.85kV, 200Ω, 25μF) using the MicroPulser electroporator, spread it on selected plates after 1h at 37℃. To test whether the editing process was controlled, we selected E. coli endogenous lacZ as a reporter gene (5’-caacagttgcgcagcctgaa-3’) for testing. The dilution of the bacterial solution under different conditions was spread on plates, containing resistant (Cm+Spec) and X-gal. The number of blue spots and white spots can be observed to determine the status of base editing.

- Fig. 1 The plasmid construction process
Next, to test the strength of these promoters, we amplified the 1500 bp promoter and 5′ UTR (5′-untranslated region) sequences of the target genes, and further constructed 20 recombinant plasmids by Gibson Assembly method. Recombinant plasmids were constructed based on the original plasmid pYLXP'-PTEF-Nluc harboring with the nanoluc luciferase gene (Nluc) as the reporter by Gibson Assembly method. For example, the recombinant plasmid of pYLXP’-P1-Nluc was constructed using linearized pYLXP'-PTEF-Nluc (digested by AvrII and NheI) and the PCR-amplified promoter P1 sequences by Gibson Assembly.
Further, to obtain the RBS library of pET plasmid rapidly, we replaced the RBS of pET-Alloferon-1-eGFP (BBa_K3868102) by referring to the NNU-China team (5'-CCGCCGGATTTACTAACTGGGGGGGGCACTAA-3'). The purpose of this step is to facilitate base editing of RBS using CBE. To enable convenient screening of optimal mutants, we refer to the high-throughput screening approach developed by Rennig et al. Specifically, we added a resistance gene (AmpR) with a stable secondary structure of SD sequence after the eGFP gene (Fig 2). When the preceding protein is expressed at a weaker intensity, the later-linked resistance genes cannot be translated properly, which plays a positive screening role. We incubated strains containing plasmids PL-CBE and RBS8G-sgRNA (BBa_K3868050) into 5mL of LB medium for 12-16h at 37°C. Then, we transferred the bacterial solution at a concentration of OD600=0.2 in 10 mL LB medium with inducer (IPTG) and associated antibiotics (Kan+Spec+Cm+Amp). We dilute and spread the bacterial solution under high concentration conditions, and the colonies were selected to 96 deep-well plates for culture. The above bacterial solution was aspirated 200μL into a 96-well plate and the unit fluorescence intensity was measured by microplate reader.
Based on the improvement of the two components, we rapidly constructed an RBS library of the pET plasmid, leading to a further 5.4-fold increase in Alloferon-1 yield.

- Fig. 2 (A) The gene after the special SD sequence cannot be produced properly at low-rate expression rate (B) The secondary structure of special SD sequences.
Result
After amplifying the 1500 bp promoter and 5′ UTR (5′-untranslated region) sequences of the target genes, we assembled these sequences with linearized pYLXP'-PTEF-Nluc (digested by AvrII and NheI) to construct the recombinant plasmids (Fig. 1 and Fig. 2), including pYLXP’-P1-Nluc(BBa_K3868110 ), pYLXP’-P2-Nluc(BBa_K3868111 ), pYLXP’-P3-Nluc(BBa_K3868112 ), pYLXP’-P4-Nluc(BBa_K3868113 ), pYLXP’-P5-Nluc(BBa_K3868114 ), pYLXP’-P6-Nluc(BBa_K3868115 ), pYLXP’-P7-Nluc(BBa_K3868116 ), pYLXP’-P8-Nluc(BBa_K3868117 ), pYLXP’-P9-Nluc(BBa_K3868118 ), pYLXP’-P10-Nluc(BBa_K3868119 ), pYLXP’-P11-Nluc(BBa_K3868120 ), pYLXP’-P12-Nluc(BBa_K3868121 ), pYLXP’-P13-Nluc(BBa_K3868122 ), pYLXP’-P14-Nluc(BBa_K3868123 ), pYLXP’-P15-Nluc(BBa_K3868124 ), pYLXP’-P16-Nluc(BBa_K3868125 ), pYLXP’-P17-Nluc(BBa_K3868126 ), pYLXP’-P18-Nluc(BBa_K3868127 ), pYLXP’-P19-Nluc(BBa_K3868128 ), and pYLXP’-P20-Nluc(BBa_K3868129 ).

- Fig. 1 The maps of recombinant plasmids for characterizing the strength of promoters
These recombinant plasmids were further transformed into Y. lipolytica po1f (Fig. 2), and obtained strains po1f-P1-Nluc, po1f-P2-Nluc, po1f-P3-Nluc, po1f-P4-Nluc, po1f-P5-Nluc, po1f-P6-Nluc, po1f-P7-Nluc, po1f-P8-Nluc, po1f-P9-Nluc, po1f-P10-Nluc, po1f-P11-Nluc, po1f-P12-Nluc, po1f-P13-Nluc, po1f-P14-Nluc, po1f-P15-Nluc, po1f-P16-Nluc, po1f-P17-Nluc, po1f-P18-Nluc, po1f-P19-Nluc, and po1f-P20-Nluc. Next, we performed the shaking flask of these engineering strains and tested the Nluc analysis.

- Fig. 2 A. The processes of constructing plasmids. B. Characterizing the strength of promoters
The results of the Nluc analysis showed that strains po1f-P2-Nluc and po1f-P17-Nluc generated the highest Nluc activities compared with other strains (Fig. 3), indicating promoter P2 and P17 are two strong promoters.

- Fig. 3 Nanoluc luciferase gene was used as a reporter gene to quantify the promoter strength.
Then, we replaced the promoter of plasmid pUC-Pexp-AnGlu(BBa_K3578010)with identified promoters P2 and P17, and obtained the recombinant plasmids pUC-P2-AnGlu and pUC-P17-AnGlu (Fig. 4), respectively. Further, we transformed plasmids pUC-P2-AnGlu and pUC-P17-AnGlu into po1f, and obtained strains po1f-P2-AnGlu and po1f-P17-AnGlu. Subsequently, we performed the shaking flask with using starch as the sole carbon source.

- Fig. 4 The maps of recombinant plasmids of pUC-P2-AnGlu and pUC-P17-AnGlu
As shown in Fig. 5, we can see that po1f-P2-AnGlu and po1f-P17-AnGlu both showed better cell growth than that of the control strain po1f-Pexp-AnGlu (BBa_K3578010), indicating that glycolase has been overexpressed by promoters P2 and P17. Moreover, among these strains, the cell growth of po1f-P2-AnGlu was the highest. That is to say that the use of strong promoter expression can improve glycolase activity, indicating that our strategy is indeed effective.
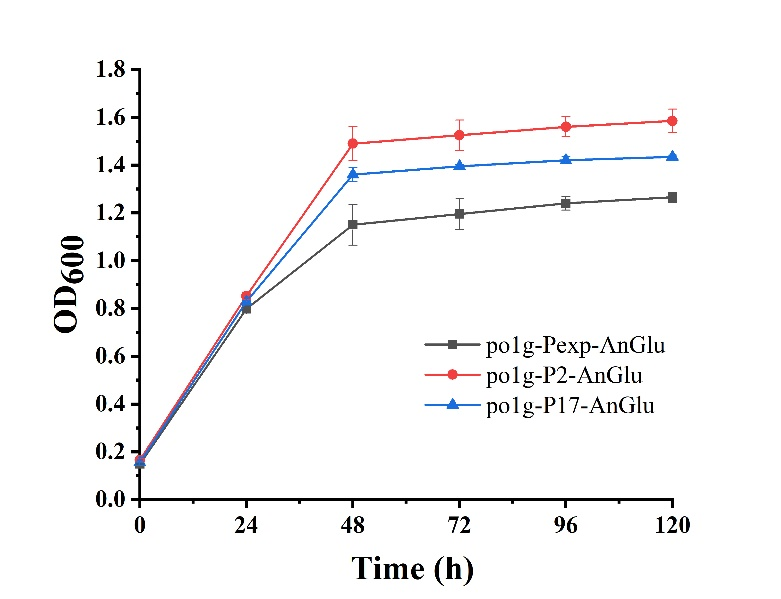
- Fig. 5 The maps of recombinant plasmids of pUC-P2-AnGlu and pUC-P17-AnGlu
References
Rennig, M., Martinez, V., Mirzadeh, K., Dunas, F., Rojsater, B., Daley, D. O., & Nørholm, M. H. (2018). TARSyn: tunable antibiotic resistance devices enabling bacterial synthetic evolution and protein production. ACS Synthetic Biology, 7(2), 432-442.
| None |
