Part:BBa_K4294559
Elowitz RBS with modified spacer region
Improvement of BBa_B0034 RBS
The context dependency of the BBa_B0034 RBS was improved by changing the biobrick spacer scar between the RBS and the ATG start codon, thus creating the RBS V59 BBa_K4294559. V59 was the result of an in silico analysis described thoroughly on our wiki page.
Original BBa_B0034 as it is distributed by iGEM: tactagag aaagaggagaaa tactaa BBa_K4294559 : tactagag aaagaggagaaa ccctcca
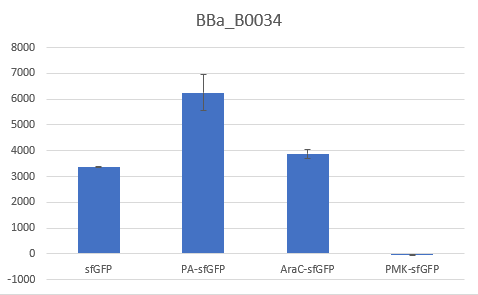
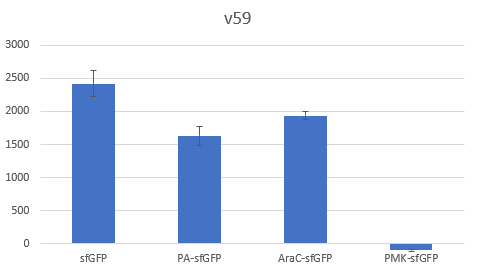
To verify the reduction in context dependence of the improved RBS spacer variants, different genetic contexts were introduced by fusions of the fluorescent protein sfGFP with the first 36 nucleotides of the proteins Penicillin acylase (PA) (BBa_K4294203), Phosphomevalonate Kinase (PMK) (BBa_K4294204) and AraC (BBa_K4294205).
The above parts together with the CDS of the sfGFP (BBa_K429420) create four genetic contexts in which the original BBA_B0034 RBS and the V59 RBS BBa_K4294559 variant was tested. Unfortunately the 36ntPMK - sfGFP fusion did not produce any fluorescence in both groups and therefore the context dependency was evaluated in three genetic contexts. The genetic circuits were engineered to be controlled by the pTet promoter and tested in DH5a-z1 cells, which have genome-encoded TetR expression. 0.5 μM of aTc was used for induction.
Bar Plot of the Fluorescence/OD (in arbitrary units) ratios of the different reporters under the translational control of the BBa_B0034 RBS after a 3h induction with 0.5 μM of aTc. The 36nt PMK - sfGFP fusion was proven not to function in both RBS groups and was excluded from the CV calculation.
Bar Plot of the Fluorescence/OD (in arbitrary units) ratios of the different reporters under the translational control of the V59 BBa_K4294559 RBS after a 3h induction with 0.5 μM of aTc. The 36nt PMK - sfGFP fusion was proven not to function in both RBS groups and was excluded from the CV calculation.
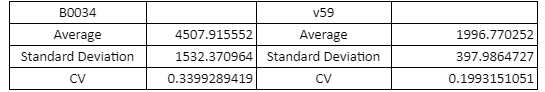
Table with Average, Standard Sample Deviation and CV values of the two RBS groups
The V59 RBS the Coefficient of Variance regarding the output signal is reduced by ~14%. The total output is also decreased; it is however in the same order of magnitude in comparison with the output of the BBa_B0034 regulated reporters.
The BBa_B0034 RBS was improved regarding its context dependence by the alteration of the RBS - Start Codon spacer region and the creation of the V59 BBa_K4294559. An obvious caveat regarding our results is the limited sample of the tested genetic contexts. However, since the choice of the potential spacer regions was supported by the in silico analysis, we believe that the V59 RBS is indeed an attractive alternative of the BBa_B0034 RBS and we encourage future iGEM teams in further characterizing it in new genetic contexts alongside with the other RBS candidate sequences that were not tested by our team. The potential reported lower translation rate could be alleviated by transcriptional enhancers leading to the reconstruction of existing RBS parts that will achieve similar average translation rates in comparison with their respective original parts, but with a lower context dependence.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |