Part:BBa_K4417023
P43-RBS-CA17-rrnB T1 Terminator in pCT5c
Description
This was the original carbonic anhydrase part from SZU-China 2017 iGEM team (Part: BBa_K2232014). We have mutated the SapI site in the coding sequence and successfully cloned into our pCT5c Type IIS compatible plasmid (BBa_K4417000).
- This part can be transformed in both B. subtilis and E. coli.
- Inducer: p-isopropyl benzoate (cumate).
- Cumate is non-toxic to the host. In our experiment, we induced the urease expression with 50 μM cumate.
- The copy number of this plasmid in B. subtilis and E. coli is unknown.
- NdeI restriction can be used to change the promoter.
Cloning Strategy
This part was cloned into pCT5c (BBa_K4417000) using restriction digest.
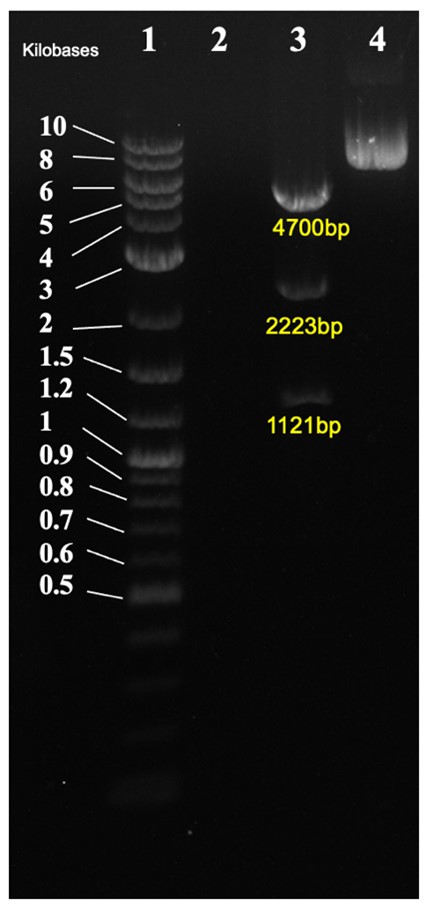
In Figure 3, the. Cloned plasmid was checked by diagnostic digest. Correct band size was observed of 4700bp, 2223bp, and 1121bp.
Characterization
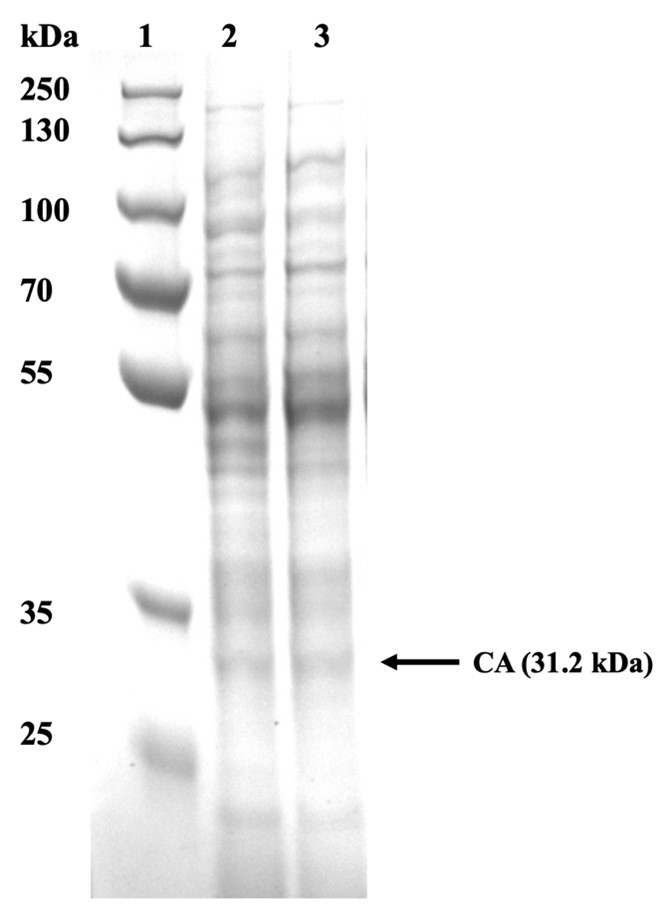
SDS Page In order to observe whether the CA22 was successfully expressed, we analysed our cell pellet using SDS PAGE. The pellet obtained from the 10 mL cultures was then resuspended in Tris Buffer Saline at an OD600. Once resuspended, the sample was cell lysed using sonication. Following sonication, the samples were spanned to separate the soluble and insoluble fragments from the whole cell lysate. 60 μL from each sample were obtained and stained with Laemmli reagent.
From Figure 4, it could be concluded that the urease was successfully produced since the correct bands are observed in cell lysate samples. Carbonic anhydrase was identified at 31.2 kDa.
Carbonic anhydrase assay We measured activity of our bacteria engineered to overexpress carbonic anhydrase (CA). We are interested in CA because it catalyzes hydration of CO2, but it can also act as an ester hydration catalyst. Combining clarified cell lysate (of cultures with OD600 = 1.74) with 4-NPA, a non-physiological commercially available ester, we were able to prove the CA activity. CA catalyzes hydration of this ester which results in yellowing of the solution. The amount of yellowing is quantified by OD348 measurements corrected for OD600 of cell cultures before lysing.
As the original assay was designed to be performed with purified protein and we didn’t have it, we decided to run it with different concentrations of clarified cell lysate to ensure proper assay sensitivity. Ultimately, we have shown that the CA17 genetic construct upregulates carbonic.
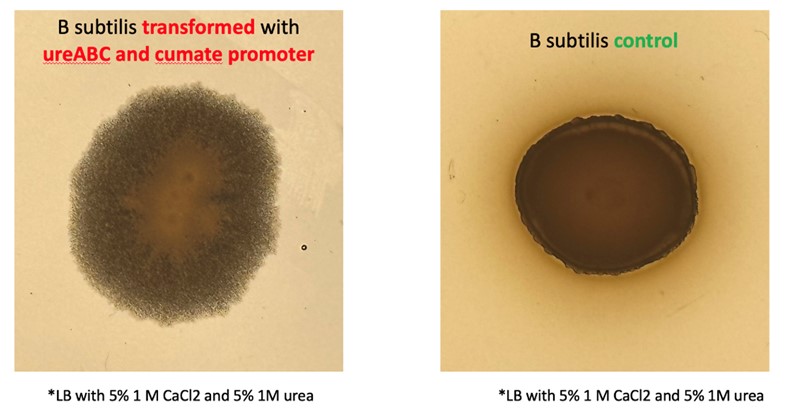
Biocementation Induced engineered bacteria should precipitate significantly more calcium carbonate in calcium rich environments than wild type bacteria. The first place where this can be observed is on the cell colony morphology. Our engineered bacteria produce crystal-like colonies (on calcium rich LB plates) which points to increased CaCO3 precipitation caused by our genetic constructs.
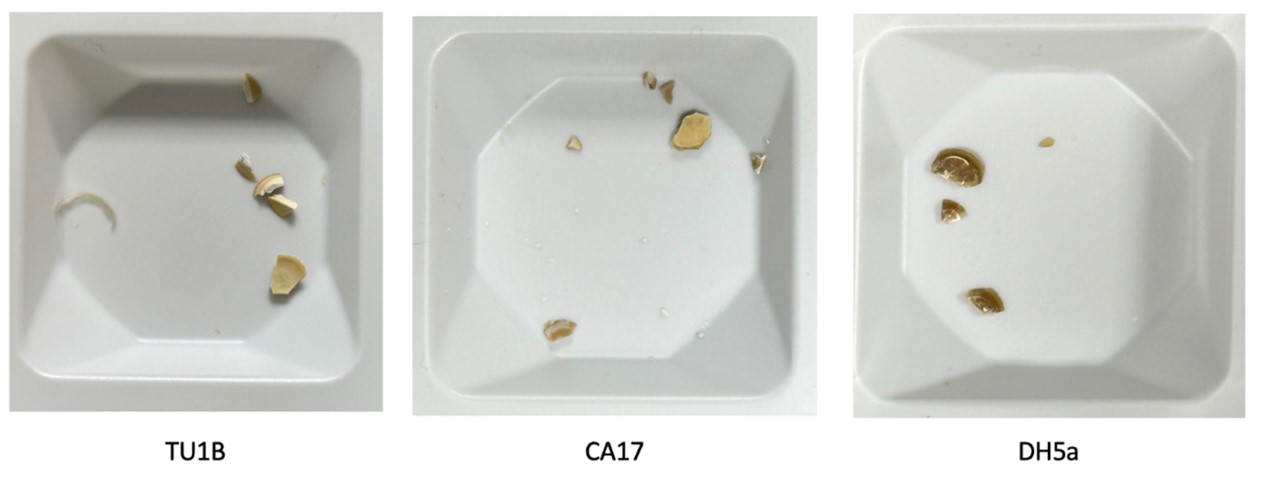
The dried aggregate of transformed bacteria is pale and matte compared with the aggregate of wild type bacteria, which is dark brown and has a glass-like surface. This is an indication of enhanced calcium carbonate precipitation.
We measured the dry weight of aggregates formed by the engineered and wild type cells in a calcium and urea rich medium. There is a statically significant (p<0.05) increase in weight between the engineered cells (TU1B, CA17) and the wild type (DH5a). The increase in dry weight of the aggregate also points to enhanced calcium carbonate precipitation of the engineered cells.
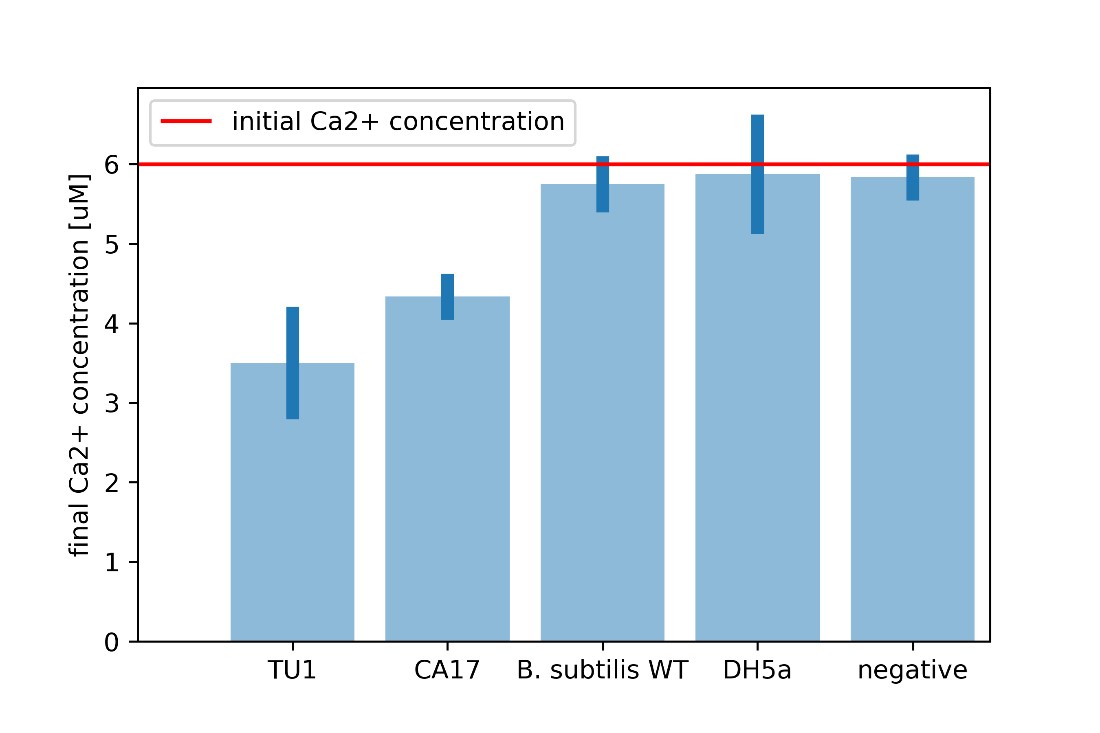
For every calcium carbonate molecule precipitated, one calcium ion leaves the solution. Calcium carbonate is insoluble in water and hence, this calcium ion can no longer interact with other chemicals in the solution. If we know the initial calcium ion concentration, we can perform a Patton-Reeder calorimetric assay to find the resulting calcium concentration. The difference between the initial and final calcium concentrations equates the calcium that left the solution either to the cell or to be bound to calcium carbonate.
It is clear that the CA17 uptakes significantly more calcium ions from the environment, combined with the observed increase in the dry aggregate weight, this proves enhanced CaCO3 precipitation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 851
Illegal NotI site found at 1138 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 831
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 376
Illegal SapI.rc site found at 534
Illegal SapI.rc site found at 846
| None |