Part:BBa_K1033001
4-coumarate ligase (4CL) with RBS
4-coumarate ligase (4CL) is an enzyme that catalyses the reaction from p-coumaric acid to 4-coumaroyl-coenzyme A (4-coumaryl-CoA). This enzyme is derived from the plant arabidhopsis thaliana, but exists in many other plants. [1]
In our project, we have been using it together with stilbene synthase, that produces resveratrol with the help of this enzyme.
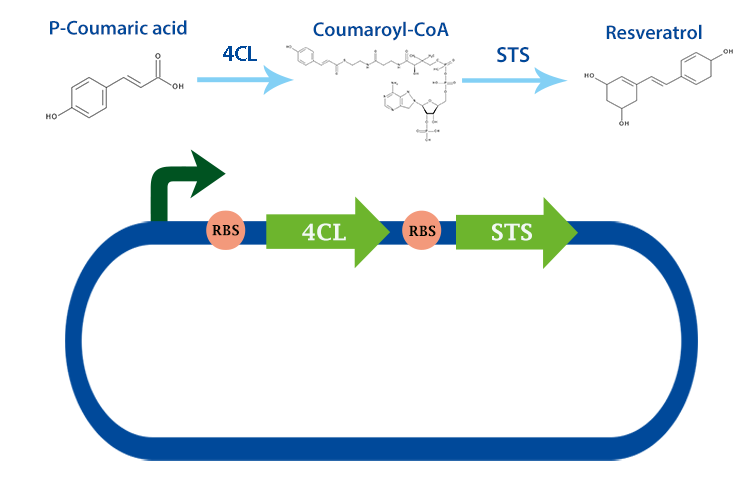
Applications With the help of this enzyme one can produce 4-coumaryl-CoA which is an important precursor in many metabolic pathways. For example, it can be used to produce resveratrol with stilbense synthase. [2]
An improved version if this part is reported on the page: https://parts.igem.org/Part:BBa_K4388002
KCL iGEM 2022
The 4-Coumarate:CoA Ligase is a member of the ANL superfamily of enzymes that includes luciferases and long-chain fatty acid CoA ligases. What unifies this superfamily is that they catalyze the adenylation of a carboxylate to form acyl-AMP followed by a second partial reaction that most often involves the formation of a thioester (Gulick, 2009).
Arabidopsis thaliana makes use of a 4CL enzyme for the synthesis of 4-coumaroyl-CoA, a precursor to a wide variety of compounds such as resveratrol or pterostilbene. 4CL is an anabolic enzyme and requires ATP to function. The stoichiometric formula is described below:
ATP + 4-coumarate + CoA = AMP + diphosphate + 4-coumaroyl-CoA
The enzyme At4CL works by accepting a carboxylate group compound and ATP in an adenylate-forming complex. Following hydrolysis of ATP, an acyladenylate intermediate is formed with the loss of two inorganic phosphates. This intermediate is then reacted with coenzyme A which forms a thioester bond, forming the many acyl sulfonamide analogues that form part of the Arabidopsis thaliana chemical repertoire (Watanebe et al., 2018).
One of these sulfonamide analogues is 4-coumaroyl-CoA. From this compound arises many pharmaceutically relevant compounds such as pterostilbene and resveratrol which have shown therapeutic potential in treating inflammatory diseases and neuroinflammation (Tomé-Carneiro et al., 2012). One noteworthy compound formed from a 4-coumaroyl-CoA precursor is the pancreatic amylase inhibitor Montbretin A which may have therapeutic potential in treating diabetes and obesity (Williams et al., 2015).
Common methods of investigating the enzymatic activity of At4CL involve a colourimetric assay. Colourimetric enzyme assays for 4-coumaroyl-CoA set colourimeters at 333 nm (Knolboch et al., 1975). Many assay kits are available online for determining enzymatic parameters of At4CL with many being designed for high throughput microplate protocols.
Subcellular Localisation
Determining the subcellular localisation of a protein using laboratory methods usually involves fluorescent protein labelling (e.g. with green fluorescent protein) coupled with fluorescence microscopy (Combs, 2010). However, these laboratory approaches have been superseded in precision by protein subcellular localisation prediction tools such as PSORTb and Proteome Analyst, and are more costly and laborious than the latter (Rey, Gardy, & Brinkman, 2005). Importantly, these computational approaches have limitations as a result of their high precision; for instance, a limitation of PSORTb is that the training datasets are limited for certain protein locations, which is mostly caused by its return of an “unknown” subcellular location if a confident prediction is not attained. Therefore, authors have emphasised the importance of complementing different computational prediction approaches to cover these pitfalls and reach a confident prediction (Gardy & Brinkman, 2006).
We took a general-to-specific approach when using different computational prediction tools to confidently characterise the At4CL mutant for its subcellular localisation, specifically regarding recombinant expression in bacteria. Two different tools, SignalP 5.0 (Armenteros et al., 2019) and PSORTb v3.0.3 (Yu et al., 2010) were used in this analysis. Firstly, SignalP detects potential signal peptides and cleavage sites across gram-negative and gram-positive bacteria, archaea, and eukarya. Then, PSORTb takes a multi-component approach whereby the amino acid sequence, motifs, signal peptides, and transmembrane helices are identified specific to 5 different bacterial locations. These were coupled to complement their distinct approaches in characterising At4CL for its subcellular localisation (Gardy & Brinkman, 2006).
Through SignalP, At4CL is predicted to not have any of the tested signal peptides from the many organism classes tested (Table 1). Further analysis for bacteria using PSORTb resulted in a confident prediction of At4CL localisation to the cytoplasm. Overall, the characterisation of the mutant At4CL for its predicted general subcellular localisation as well as for recombinant expression in microorganisms such as E. coli has provided a strong prediction of its localisation. Beyond characterisation itself, this also suggests that the expressed At4CL mutant is correctly located within the bacteria for pterostilbene production.
Table 1. SignalP Prediction for Probability of Signal Peptides in Different Organisms Values depict the probability (0 to 1) of the input amino acid sequence of the At4CL mutant to contain any of three types of signal peptides: Sec/SPI, Sec/SPII, and Tat/SPI, across Eukarya, Gram-positive and Gram-negative bacteria, and Archaea. Results are given to 3 decimal places.
Citations
Almagro Armenteros, J. J., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., . . . Nielsen, H. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology, 37(4), 420-423. doi:10.1038/s41587-019-0036-z
Gulick A. M. (2009). Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS chemical biology, 4(10), 811–827. https://doi.org/10.1021/cb900156h
Tomé-Carneiro, J., Gonzálvez, M., Larrosa, M., Yáñez-Gascón, M. J., García-Almagro, F. J., Ruiz-Ros, J. A., García-Conesa, M. T., Tomás-Barberán, F. A., & Espín, J. C. (2012). One-year consumption of a grape nutraceutical containing resveratrol improves the inflammatory and fibrinolytic status of patients in primary prevention of cardiovascular disease. The American journal of cardiology, 110(3), 356–363. https://doi.org/10.1016/j.amjcard.2012.03.030
Williams, L. K., Zhang, X., Caner, S., Tysoe, C., Nguyen, N. T., Wicki, J., . . . Brayer, G. D. (2015). The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif. Nature Chemical Biology, 11(9), 691-696. doi:10.1038/nchembio.1865
Knobloch, K.-H. et Hahlbrock, K.. (1975). Isoenzymes of p-Coumarate: CoA Ligase from Cell Suspension Cultures of Glycine max. European journal of biochemistry, 52(2), 311‑320. doi:10.1111/j.1432-1033.1975.tb03999.x
Watanabe, B., Kirikae, H., Koeduka, T., Takeuchi, Y., Asai, T., Naito, Y., . . . Hiratake, J. (2018). Synthesis and inhibitory activity of mechanism-based 4-coumaroyl-CoA ligase inhibitors. Bioorganic & Medicinal Chemistry, 26(9), 2466-2474. doi:https://doi.org/10.1016/j.bmc.2018.04.006
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1108
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1675
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1307
This part has been improved upon, as detailed below. The new part BBak4388002 has been shown by Yan et. al (2021) to have improved catalytic efficiency, and we have further characterised it. BBa_K1033001 has the same protein sequence as BBa_K801093.
Molecular Docking by KCL_UK
Introduction
As part of engineering our plasmid constructs to include the genes for four enzymes in the pathway to synthesize pterostilbene, ensuring the most efficient enzymes possible was important. The four genes we decided to use for our biosynthetic pathway for pterostilbene production were mutant versions of the wild type forms of the enzymes RgTAL, At4Cl, VvSTS and VvROMT found by (Yan et. al, 2021). These mutations claimed to increase the pterostilbene production titre by a factor of 13.7 compared to their respective wild type forms. As this specific mutant At4CL1 was found in literature to have greater catalytic efficiency than the Wild-type (Yan et. al, 2021), it presented a potential variant to use in our plasmid constructs. As the wild type form of At4CL was already present in the iGEM registry under the code BBa_K1033001, created by iGEM13_Uppsala, we decided to explore experimental avenues that would support and further characterize this wild type and mutant variant (BBak4388002) to justify our reasoning for choosing the mutant form of the enzymes and to improve the part. We aimed to determine differences in binding energy and affinity between this mutant and its wild type computationally to further investigate and characterise the mutant and wild type for use in our project.
At4CL
4-Coumaroyl-CoA Ligase (4CL) catalyses the conversion of p-Coumaric acid to p-Coumaroyl-CoA. The BioBrick At4CL1 part BBa_K1033001 has 100% local identity with Genbank Accession number AAA82888.1, and Uniprot accession code Q42524.
The mutant At4CL1 found to have greater catalytic efficiency than the Wild-type has point mutations at L57I and L460H.(Yan et. al, 2021) Docking simulations were performed to obtain KD and ΔG of both the wild-type and mutant.
Obtaining PDB models for the Wild Type and Mutant
The closest template found from the Swiss Model was part of a fusion protein with Stilbene Synthase from (Wang et. al, 2011). Structural differences of At4CL1 as part of the fusion protein were reported to not vary drastically when compared with At4CL1 alone,(Wang et. al, 2011) and the Root Mean Squared Deviation (RMSD) value comparing the 4CL section of the 3TSY fusion protein to the At4CL1 Alphafold model was low (0.445), suggesting the Alphafold model is similar in structure with the 3TSY model, as can be seen in Figure 1. As we were investigating whether point mutations in only two locations had an effect on binding energy, obtaining the most accurate PDB model was important. Using a model of a homolog would not have been accurate enough for docking simulations, given that catalytic specificity and efficiency for a specific substrate can vary significantly even between the different At4CL isoforms. The Alphafold model was chosen as it was of high confidence, and had a more complete structure than 3TSY could provide.(Figure 1 and 2) The mutant At4CL1 PDB was created by mutagenesis in Pymol of the Wild-type Alphafold model to introduce the mutations L57I and L460H. Our new part, BBak4388002, contains these mutations.
 "
"
Figure 1. Alphafold model (purple), 4CL section from fusion protein 3TSY (blue). RMSD = 0.445. Made using Pymol. The Alphafold structure can be seen to be more complete than the 4CL section from the fusion protein 3TSY.

Choosing a Docking programme
Predicted docking energies from both Autodock Vina or Autodock4 have been found to correlate well with experimentally determined docking energies, with values obtained using Autodock4 consistently closer to experimentally determined values. Autodock4 has been found to be the superior option for estimating binding affinity (Nguyen et. al, 2020), making it the preferred option for our purposes. Yasara Structure was therefore used to perform docking simulations using Autodock4.
Using Yasara
Energy minimisation was run in Yasara for both enzyme and substrate to find the most energetically favourable conformations. The PDB structures for the enzymes were found to have improved Molprobity results after Energy minimisation compared to before. Each Autodock4 docking simulation performed 25 runs, and clustered results with high similarity into distinct complex conformations. The results from the best-scoring distinct complex conformation for wild type and mutant according to Autodock4 was selected for comparison.
Results and Conclusion
The value of KD found for the mutant At4CL1 was lower than that of the wild type, suggesting it has better affinity with p-Coumaric acid. (Table 1) This may help explain the greater catalytic efficiencies of the mutant variant compared to its wild type.(Yan et. al, 2021) The At4CL mutant had a more favorable binding energy change with p-Coumaric acid than the wild-type.


Figure 3. Yasara docking simulation result with Autodock4. Shown is the Wild-type 4-Coumaroyl-CoA Ligase enzyme from Arabidopsis thaliana in complex with the substrate p-Coumaric acid. Yasara automatically colour codes the secondary structure elements as follows: Alpha helices (dark blue), inside of helix (grey), beta sheets (red), turn (light green), helix 310 (yellow), coil (light blue).
From the Yasara docking simulation, the results for wild type and mutant were compared in Pymol. The position of p-Coumaric acid when bound to wild type and mutant At4CL is different and can be seen in figure 4.

Figure 4. p-Coumaric acid (Yellow) in complex with wild-type At4CL (Light blue), overlaid onto p-Coumaric acid
(Red) in complex with mutant At4CL (Dark blue). Created in Pymol using Yasara docking simulation results.
Further examination of the amino acid residues of At4CL involved in binding to p-Coumaric acid can be seen in Figure 5. As seen in Table 2, though most of the amino acid residues involved in binding to p-Coumaric acid are predicted to be the same in both mutant and wild type, some residues involved in the binding are different in the mutant and wild-type.
Table 2. At4CL amino acid residues involved in binding to p-Coumaric acid. Comparisons of amino acid residues from wild type and mutant At4CL from Autodock4 results using Yasara


Figure 5. A) p-Coumaric acid (purple) in complex with the wild-type At4CL. B) p-Coumaric acid (Dark green) in complex with mutant At4CL. Created in Pymol.
References
Wang, Y., Yi, H., Wang, M., Yu, O., & Jez, J. M. (2011). Structural and kinetic analysis of the unnatural fusion protein 4-coumaroyl-CoA ligase::stilbene synthase. Journal of the American Chemical Society, 133(51), 20684–20687. https://doi.org/10.1021/ja2085993
Nguyen, N. T., Nguyen, T. H., Pham, T., Huy, N. T., Bay, M. V., Pham, M. Q., Nam, P. C., Vu, V. V., & Ngo, S. T. (2020). Autodock Vina Adopts More Accurate Binding Poses but Autodock4 Forms Better Binding Affinity. Journal of chemical information and modeling, 60(1), 204–211.. https://doi.org/10.1021/acs.jcim.9b00778
Yan, Z. B., Liang, J. L., Niu, F. X., Shen, Y. P., & Liu, J. Z. (2021). Enhanced Production of Pterostilbene in Escherichia coli Through Directed Evolution and Host Strain Engineering. Frontiers in microbiology, 12, 710405. https://doi.org/10.3389/fmicb.2021.710405
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., Potapenko, A., Bridgland, A., Meyer, C., Kohl, S., Ballard, A. J., Cowie, A., Romera-Paredes, B., Nikolov, S., Jain, R., Adler, J., Back, T., … Hassabis, D. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873), 583–589. https://doi.org/10.1038/s41586-021-03819-2
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., Yuan, D., Stroe, O., Wood, G., Laydon, A., Žídek, A., Green, T., Tunyasuvunakool, K., Petersen, S., Jumper, J., Clancy, E., Green, R., Vora, A., Lutfi, M., Figurnov, M., … Velankar, S. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic acids research, 50(D1), D439–D444. https://doi.org/10.1093/nar/gkab1061
Krieger, E., & Vriend, G. (2014). YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics (Oxford, England), 30(20), 2981–2982. https://doi.org/10.1093/bioinformatics/btu426
The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
iGEM 2022 SHSBNU-China, new documentation (For Bronze)
Group: SHSBNU-China iGEM 2022
In this year’s iGEM competition, SHSBNU_China team focused on anthocyanins production, especially delphinidin. 4CL gene is one of the key genes in the synthetic pathway of delphinidin, which catalyzed phenylalanine into 4-coumaryl coenzyme A.
We synthesized the sequence and constructed into plasmid pETDuet.
After transforming to E.coli BL21 and induced by IPTG overnight at 16 degree Celsius at various concentrations (0.25 to 2 uM), we lysed the bacteria and performed SDS-PAGE and we confirmed 4CL enzyme to be expressed successfully. Judged by the thickness of the SDS-PAGE bandwidth and darkness, concentrations of iPTG don’t play a critical role in the expression of 4CL enzyme.
//collections/probiotics/production
| None |


