Part:BBa_K4130000
Contents
IgG F(c) Binding Protein, EibD
Biology
Escherichia coli Ig-binding proteins (Eibs) are a wide-spread class of proteins within intimin-negative, shiga-toxin producing enterohemorrhagic E. coli (EHEC) strains [1]. First discovered in strain ECOR-9 [2, 3], the class has grown to include EibA, C, D, E, F, and G. The class is united in its common characteristic of immunoglobulin (Ig) binding activity.
Immunoglobulins, also known as antibodies, are a family of heterodimeric proteins produced by B cells capable of binding with great specificity to respective antigens. Igs are typically composed of a “variable region”, responsible for binding of the antigen, and a “constant region” with no affinity for the antigen. Igs are further classified according to their “constant regions”, into 5 classes: IgM, IgG, IgA, IgD, and IgE. [4]
E. coli Ig-binding proteins have been shown to be capable of binding the constant region of assorted Ig classes. Affinity and specificity for different classes of Ig proteins varies between Eib family members, with dissociation constants on the 100 nM scale [5].
EibD (from E. coli strain ECOR-9) is a 511 amino acid, 210 kDa [3] member of the Eib family. The protein exhibits affinity toward both species-nonspecific IgG (Kd = ~73 nM) and IgA (Kd = ~135 nM) [5], with separate binding domains for each Ig [6]. The structure also includes a membrane anchor, left-handed coiled-coil, saddle domain, right-handed coiled-coil, neck, and head domain [6], characteristic of trimeric autotransporter adhesins - a family of secreted proteins in Gram-negative bacteria that are associated with virulence [7]. In addition to Ig-binding properties, heterologous expression of EibD has resulted in whole-cell self-aggregation phenotypes [6], typical of other trimeric autotransporter adhesins such as YadA [7]. Transmission electron microscopy (TEM) imaging has revealed zipper-like structures forming between neighboring bacteria harboring EibD, explaining the autoaggregation phenotype. Although the biological function of EibD is unknown, its Ig-binding properties and homology to other autotransporter adhesins implicates its role in serum resistance; binding of IgG by EibD may block binding of immunoglobulins to the adaptive immunity protein C1q, protecting bacteria against innate host defenses [6].
Usage
A major avenue of synthetic biology research is the development of whole-cell bacterial biosensors: bacteria that have been genetically engineered to “sense” and “report” the concentration of molecules of interest. These biosensors represent a promising improvement to current biosensors, due to their low-cost, self-manufacturing, and biodegradable properties. However, advancements in whole-cell bacterial detection remain hindered by difficulties associated with having to re-engineer a new solution for each compound of interest. In response to this challenge, we have designed an EibD-based universal biosensor. We chose EibD for its Ig (antibody)-binding properties. Expression of EibD results in auto-agglutination and biofilm formation that can be observed via the eye [6]. By incubating EibD-expressing bacteria with immunoglobulin antibodies, we hoped to be able to detect various antigens. Introduction of the antibodies would cause visual and quantitative dis-agglutination by molecular competition between self-self interactions and self-antibody interactions. Addition of the antigen would lead to multivalent binding interactions (multiple antibody-bound bacteria binding the same antigen), causing re-agglutination of the cells. (Figure 1)
Accordingly, this strategy of detection has the potential to be applied to nearly any antigen with zero need for further genetic engineering. Previous whole-cell bacterial sensors have been focused on expressing antibodies or antibody fragments on the surface of the bacterium, meaning that each strain is specific to an antigen. By expressing a general antibody-binding protein, a single strain can be utilized for the detection of multiple antigens.
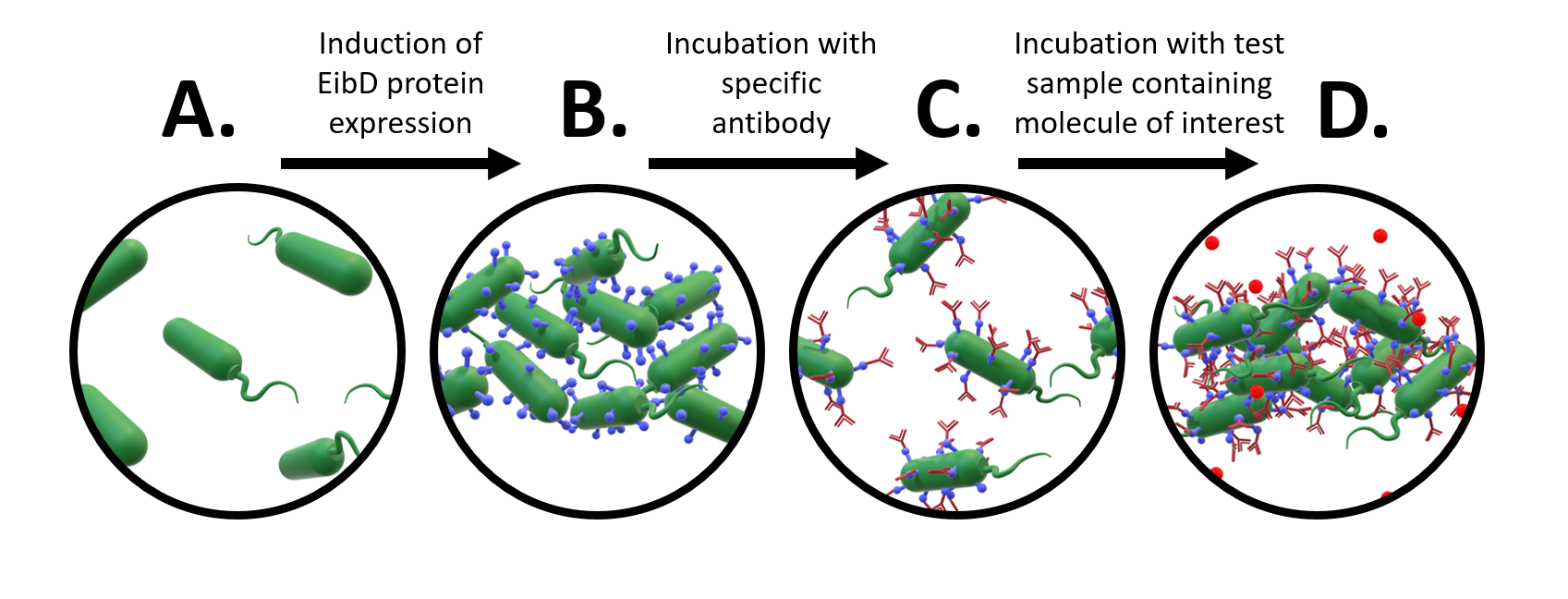
Figure 1: Proposed Design of Universal Whole-Cell Biosensor. a) uninduced E. coli exhibit relatively few interactions b) after induction of EibD expression, bacteria begin to "autoaggregate", forming clumps due to homophilic EibD-EibD interactions c) incubation with an antibody reduces bacterial autoaggregation due to competition between EibD-antibody and EibD-EibD binding interactions d) after incubation with a sample containing the antigen, multivalent interactions form, where multiple antibodies bind to the same antigen. This results in an increase in autoaggregation.
In keeping with the overarching goal of our project, we chose to exploit the design of our universal whole-cell biosensor for the detection of the small molecule asparagine. Unlike larger molecules such as proteins, asparagine presents a unique challenge in that it contains limited theoretical binding sites for antibodies. We therefore had to make modifications to the design of our biosensor to accommodate monovalent antibody-antigen interactions. In this modified design, a non-antigenic protein is crosslinked to multiple small molecules of interest, forming an antigenic "bead". This "bead" can be used to induce multivalent antibody-antigen interactions, similar to what happens in the case of a larger molecule of interest. These multivalent interactions can subsequently be competed out by free antigen present in a test sample. (Figure 2)

Figure 2: Application of EibD whole-cell biosensor to detecting amounts of free asparagine in maple syrup. A) Uninduced E. coli B) EibD-expressing bacteria exhibiting autoaggregating phenotype C) Reduction in autoaggregating phenotype after incubation with anti-asparagine antibody D) Addition of BSA-Asparagine beads that allow for bivalent interactions between the antibodies, causing agglutination (re-clumping) of the bacteria E) Addition of "test sample" containing free asparagine causes molecular competition for binding free asparagine and bead-bound asparagine. This results in a reduction in agglutination proportional to the concentration of free asparagine competitor.
We therefore designed a genetic circuit composed of the EibD gene from E. coli strain ECOR-9, a rhamnose-inducible promoter (BBa_K914003), strong ribosome binding site (BBa_B0034), and double-terminator (BBa_B0015). Synthesized parts were assembled into the chloramphenicol-resistance conferring pSB1C3 vector.
Characterization
The EibD biobrick was assembled into the pSB1C3 vector by 3A assembly and cloned into E. coli DH5a for storage and E. coli BL21 for expression experiments.
Induction of EibD Expression Results in Whole-Cell Autoaggregation Phenotype
Cultures of E. coli BL21 (+/- biobrick BBa_K4130000) were grown to mid-exponential phase (O.D.600 ~ 0.4 - 0.6), where growth and protein translation is maximal. Subsequently, to determine the differential effect of inducer concentration, bacteria were induced with 0.0% rhamnose, 0.001% rhamnose, or 0.01% rhamnose and grown for 2 hours at 30 C, 200rpm. After induction, the EibD-transformed bacterial cultures appeared markedly different from a BL21 control strain harboring no plasmid. A white precipitate accumulated on the bottom of the EibD cultures induced with 0.001% L-rhamnose, while the uninduced EibD culture exhibited no formation of white precipitate (Figure 3). This white precipitate is likely a result of autoaggregation due to previously reported homophilic interactions between EibD proteins on separate bacteria [6]. To investigate this, we used light microscopy to visualize the bacteria. Bacterial cultures were pelleted and washed in PBS, followed by incubation with acidic proteoglycan-staining safranin (Figure 4). Control E. coli BL21 without the EibD-containing plasmid appear evenly dispersed throughout the field of view. In contrast, EibD-expressing E. coli BL21 appear to aggregate in clumps within the field of view. This evidence is further indicative of autoaggregation. We quantified the degree of autoaggregation by measuring the settling rate of bacteria in solution over time. After brief mixing, the optical density at 600 nm (O.D.600) was recorded over a period of 30 minutes. As the bacteria settled and precipitated over time, a decrease in optical density was observed. The O.D.600 was observed to decrease more rapidly with increasing concentrations of L-rhamnose inducer, indicating that autoaggregation correlates positively with induction concentration. (Figure 5)

Figure 3: Visual Observation of Autoaggregation Mid-exponential phase BL21 cultures (with and without EibD-harboring plasmid) were induced with 0% or 0.001% L-rhamnose and grown for 2 hours. 2mL of pelleted bacteria was resuspended in PBS, briefly mixed, and allowed to settle over a period of 70 minutes. Images were taken at 0 minutes, 40 minutes, and 70 minutes.
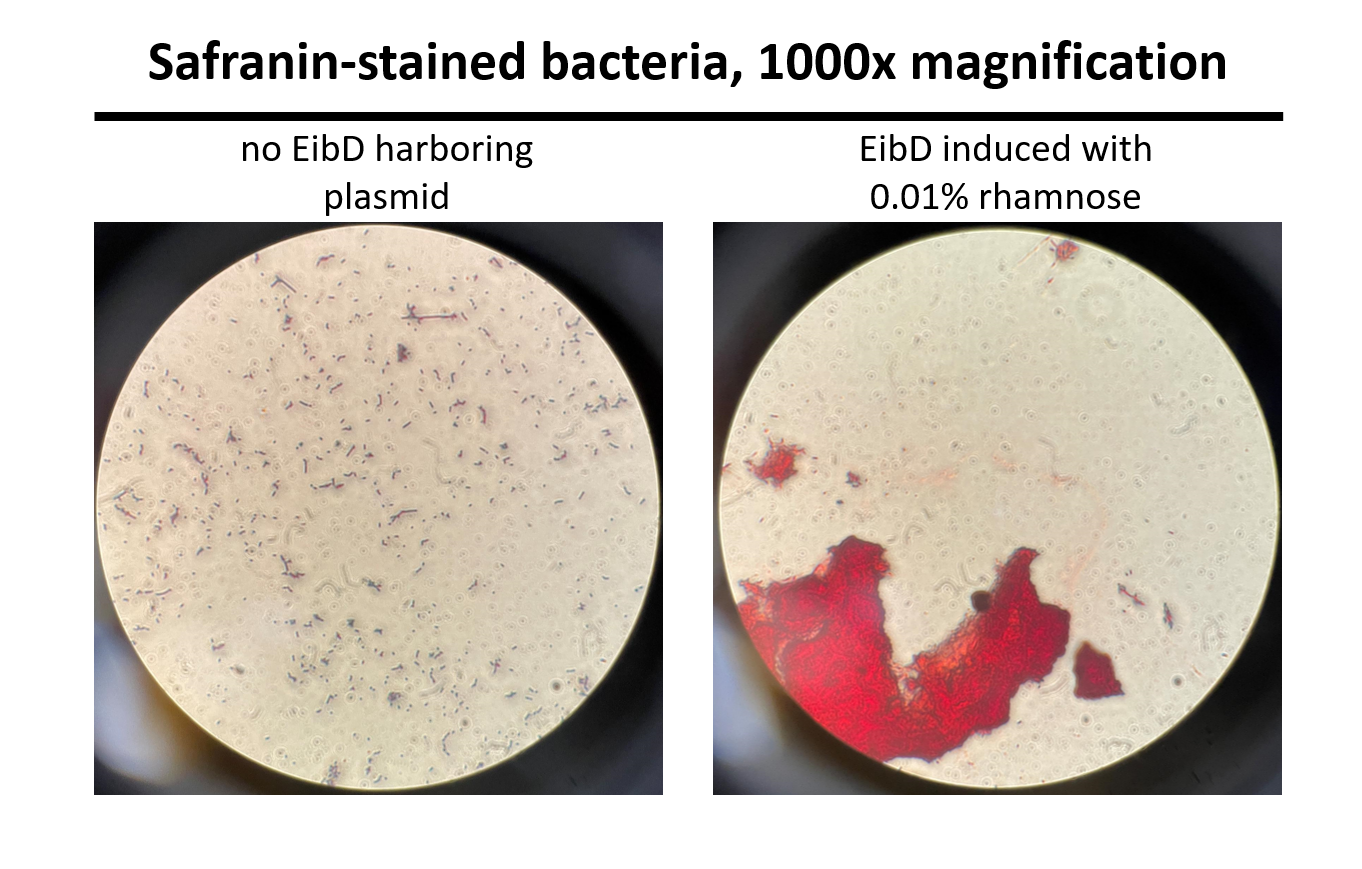
Figure 4: Light Microscopy Observation of Autoaggregation Induced bacteria were stained with safranin and visualized using a light microscope at 1000x magnification. Pictures were taken through the eyepiece using a phone camera. As can be seen on the left, control E. coli BL21 bacteria are evenly distributed throughout the field of view. On the right, EibD-induced bacteria appear to clump together, indicative of autoaggregation.
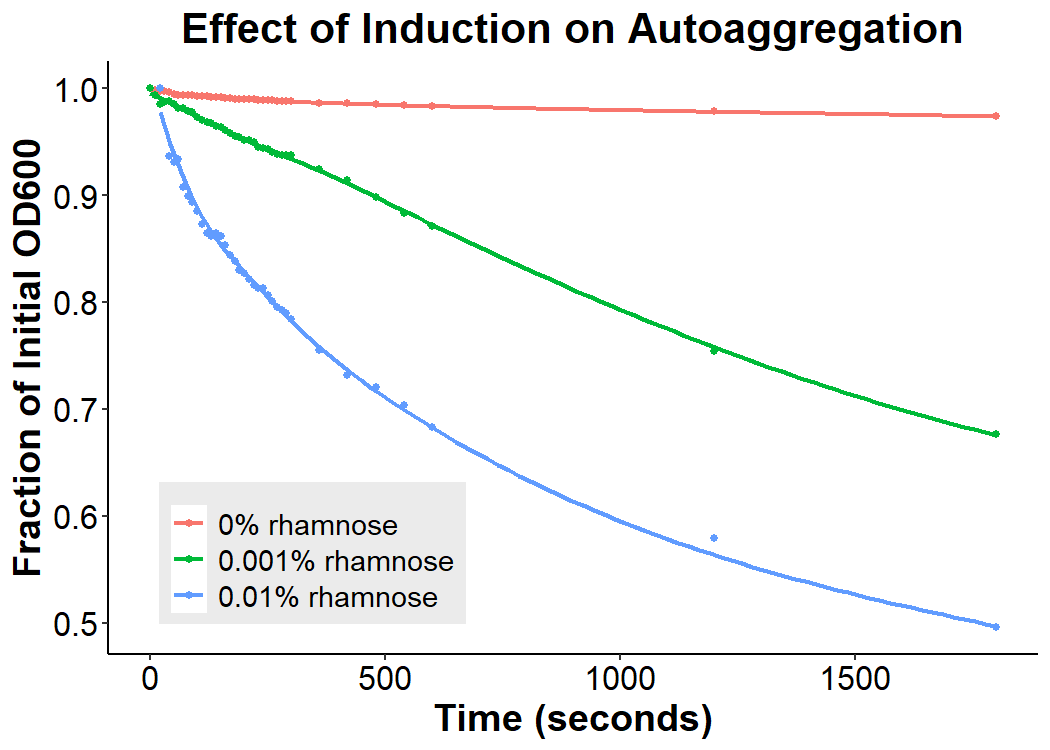
Figure 5: Effect of Induction Concentration on Agglutination Mid-exponential phase BL21 EibD-transformed cultures were induced with 0%, 0.001%, and 0.01% L-rhamnose and grown for 2 hours. 2mL of pelleted bacteria was resuspended in PBS, briefly mixed, and the optical density at 600nm was monitored for 30 minutes.
EibD Expressed on Live Cells Binds FITC-Tagged IgG
EibD has previously been shown to be capable of non-immune binding to IgG [10]. The premise of our biosensor depends heavily upon this capability. Therefore, it was necessary to demonstrate that expression of BioBrick BBa_K4130000 in E. coli BL21 produces functional EibD capable of binding IgG. The EibD-harboring strain was induced with 0%, 0.001%, or 0.01% L-rhamnose and cultured for 2 hours. Bacteria were harvested and washed in PBS two times. Subsequently, bacteria were incubated in 200ug/mL mouse IgG-FITC for 24 hours. Bacteria were pelleted and washed twice, followed by fixation and staining for DNA by NucBlue. (Figure 4).
Comparing Figure 6A-C, as inducer concentration is increased, large clumps of bacteria become apparent. This is suggestive of autoaggregation, and further supports the quantitative data in Figure 5, and qualitative data in Figures 3 and 4.
Comparing Figure 6A and D, B and E, and C and F indicates localization of the IgG-FITC protein and the DNA. Throughout all induction levels, IgG-FITC signal is weak, except for exceedingly bright spots that do not colocalize with the DNA. These bright spots likely represent aggregates of the IgG-FITC protein in solution. The weaker “blots” of FITC fluorescence do, however, co-localize with the DNA stain in Figure 6, panels A-C. This suggests that IgG-FITC may be interacting with the EibD-expressing bacteria as observed in previous research.
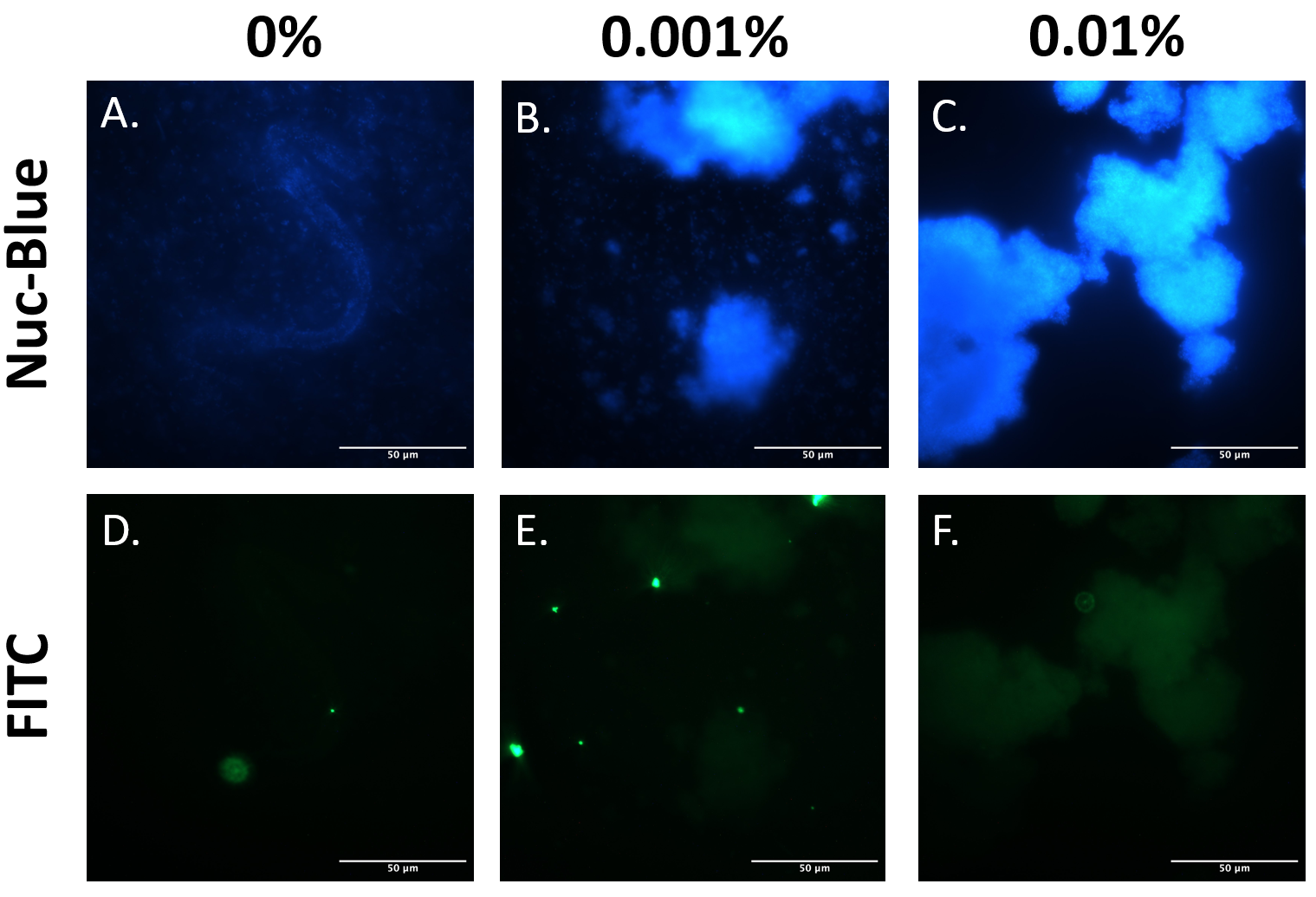
Figure 6: IgG-FITC Co-Localizes With EibD-Induced Bacteria Harvested bacterial cultures induced with 0%, 0.001%, and 0.01% rhamnose were incubated with mouse IgG-FITC for 24 hours and then washed. Fixed bacteria were stained for DNA using NucBlue Fixed Cell ReadyProbes Reagent and imaged. Panels A and D, B and E, and C and F correspond to the same region. .
References
Merkel V, Ohder B, Bielaszewska M, Zhang W, Fruth A, Menge C, Borrmann E, Middendorf B, Müthing J, Karch H, Mellmann A. Distribution and phylogeny of immunoglobulin-binding protein G in Shiga toxin-producing Escherichia coli and its association with adherence phenotypes. Infect Immun. 2010 Aug;78(8):3625-36. doi: 10.1128/IAI.00006-10. Epub 2010 Jun 14. PMID: 20547747; PMCID: PMC2916290.
Sandt, C. H., Y. D. Wang, R. A. Wilson, and C. W. Hill. 1997. Escherichia coli strains with nonimmune immunoglobulin-binding activity. Infect. Immun. 65:4572-4579.
Sandt, C. H., and C. W. Hill. 2000. Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Escherichia coli. Infect. Immun. 68:2205-2214.
Schroeder HW Jr, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010 Feb;125(2 Suppl 2):S41-52. doi: 10.1016/j.jaci.2009.09.046. PMID: 20176268; PMCID: PMC3670108.
Leo, J. C., Goldman, A. 2009. The immunoglobulin-binding Eib proteins from Escheria coli are receptors for IgG Fc. Molecular Immunology 46(8-9):1860-1866.
Leo, J. C., Lyskowski, A., Hattula, K., Hartmann, M. D., Schwarz, H., Butcher, S. J., Linke, D., Lupus, A. N., Goldman, A. 2011. The Structure of E. coli IgG-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins. Structure 19(7):1021-1030.
D. Linke, T. Riess, I.B. Autenrieth, A. Lupas, V.A. Kempf. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol., 14 (2006), pp. 264-270
| None |
