Part:BBa_K4345001
RNA Thermometer Hsp17
Hsp17 is in nature implemented in the mRNA structure of a cyanobacterial (Synechocystis sp.) heatshock gene which is important for stress tolerance. The thermometer is a structural element that is located within the 5’ UTR of protein-coding mRNA. It operates as a reversible molecular zipper that controls the availability of the RBS in its structure. When the temperature increases, the thermometer unzips and the RBS can be detected by the ribosome thus allowing translation of downstream mRNA. When the temperature is below a certain range, the RBS gets trapped (Kortmann et al., 2010).
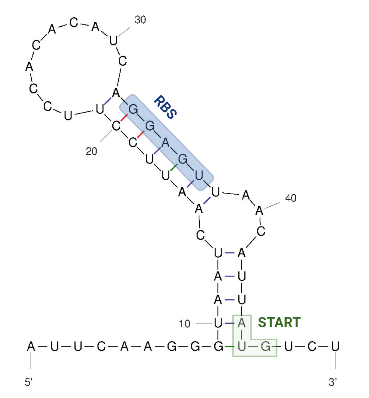
The 5’ UTR end consists of a single hairpin with an internal asymmetric loop, as seen in the figure below. The secondary structure was predicted by RNAFold. The trapped RBS sequence is highlighted in blue. The start codon is highlighted in green and is located 45 nucleotides downstream from the transcriptional start site which means that the thermometer is very short compared to others that are described.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
The figure below shows the secondary structure of the RNA thermometer.
Image obtained from Kortmann et al., 2010, edited with biorender and UNAfold.
Results
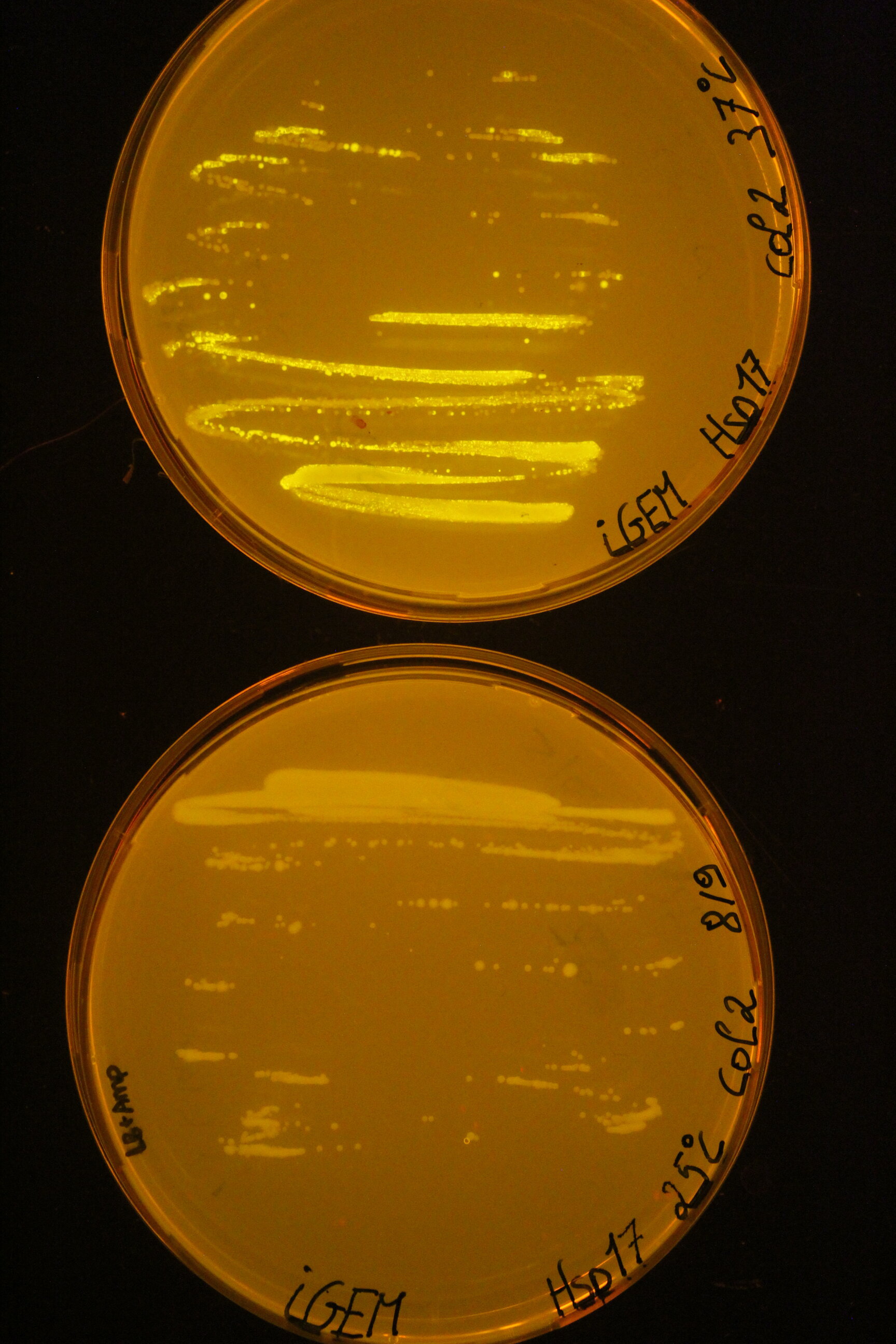
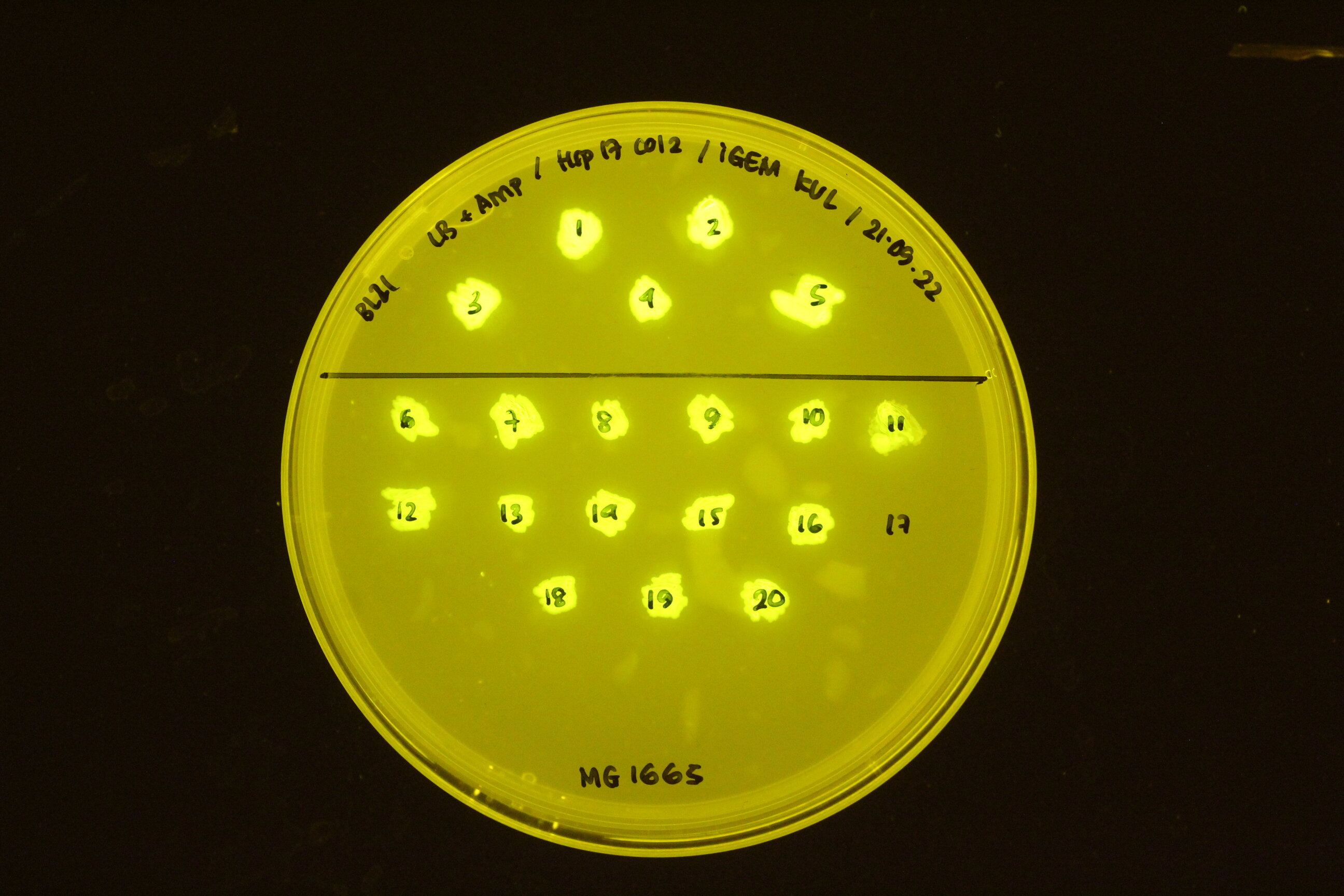
Below you can find an example of the results found after the induction tests of the RNA thermometer.
Two 96 well plates were prepared with 200 µL of the relevant selection medium (LB + 100 µg/mL ampicillin) and the plates were covered with a breathable film. The cultures E. coli DH5α were diluted 50 times in these well plates and grown at 25°C or 37°C overnight at 180 rpm. The next day, both the OD600 as the fluorescence intensity were measured with the CLARIOstar (BMG Labtech, Ortenberg, Germany). Blancs (LB) and negative controls (culture with pLO_SNAP plasmid) are subjected to the same protocol. All measurements were performed in triplicate. To visualise the results for this picture, the successful cultures were grown on plates at the different temperatures.
In the figure below, the construct transformed into different cell types: BL21 (top) and K12 M1665 (bottom).
References
Kortmann, J., Sczodrok, S., Rinnenthal, J., Schwalbe, H., & Narberhaus, F. (2010, December 3). Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Research, 39(7), 2855–2868. https://doi.org/10.1093/nar/gkq1252
//rna/riboswitch
| biology | |
| control |