Part:BBa_K4247007
Contents
- 1 Minispidroin_NT-2rep-CT_N-6His
- 2 Usage and Biology
- 3 Characterization
- 3.1 Optimization of inducer concentration
- 3.2 Optimization of temperature after induction
- 3.3 Optimization of lysis buffer
- 3.4 Heat purification
- 3.5 pH purification
- 3.6 Optimization of media
- 3.7 Protein purification by IMAC
- 3.8 A quicker approach to protein purification by IMAC
- 3.9 Stripping the Ni-NTA columns with urea
- 3.10 Large scale production
- 3.11 Spinning
- 3.12 Protein yields
Minispidroin_NT-2rep-CT_N-6His
This part codes for the full minispidroin protein, a highly soluble spider silk protein, with a 6x His-tag in the N-terminus. This is a composite part consisting of the following basic parts: BBa_K4247005 (Minispidroin_NT_N-6His), BBa_K4247001 (Minispidroin_2rep) and BBa_K4247002 (Minispidroin_CT). BBa_K4247007 contains the coding sequence for the full minispidroin protein with 2 repeats of the central repetitive domain with a 6x His-tag in the N-terminus of the protein.
This part is one of a collection of compatible minispidroin parts: BBa_K4247000 (Minispidroin_NT), BBa_K4247001 (Minispidroin_2rep), BBa_K4247002 (Minispidroin_CT), BBa_K4247004 (Minispidroin_NT-2rep-CT), BBa_K247005 (Minispidroin_NT_N-6His), BBa_K247007 (Minispidroin_NT-2rep-CT_N-6His), BBa_K247010 (Minispidroin_NT-2rep-CT-SnoopTag_N-6His), BBa_K247011 (Minispidroin_NT-4rep-CT), BBa_K247012 (Minispidroin_NT-4rep-CT_N-6His), BBa_K247013 (Minispidroin_NT-4rep-CT-SnoopTag_N-6His).
Usage and Biology
Dragline silk produced by spiders is one of the strongest natural materials to exist and it is mainly made up of structural proteins called spidroins. These spidroins consist of non-repetitive N-terminal and C-terminal domains and a repetitive central part consisting of tandem repeats of a certain amino acid sequence. These sequences are rich in alanine and glycine to form the crystalline and amorphous parts of the fibre respectively.
There are many research articles whose authors could successfully produce recombinant spider silk proteins and spin them into fibres by mimicking the conditions of the spider’s silk gland where the fibers are formed naturally. But a major drawback in many of these recombinant spidroins was their low solubility. It has been found that the N-terminus of the spidroin is highly soluble at neutral pH which contributes to the solubility of the protein.
In the spider's silk gland, before spinning, the spidroins remain in a highly concentrated and soluble state. Then, this highly concentrated spidroin solution called spinning dope is subject to a gradual drop in pH from 7.6 to 5.7 along the gland which triggers the formation of the fiber. This drop in pH triggers the N-terminus to be more stable and form large network-like structures whereas the C-terminus becomes more unstable to drive spontaneous fibre formation by forming the beta-sheet fibrils which form the core of the fiber. The N-terminal domain restricts the formation of silk fibers to a precise point in the silk duct, preventing silk proteins stored in the silk gland from agglutinating.
This clearly shows us that the solubility and pH sensitivity have a huge effect on the N- and C-terminus of the spidroin which thus affects the formation of fibers. It has been found that the N-terminus of MaSp1 (Major ampullate spidroin 1) from Euprosthenops australis, shows extremely high solubility and pH sensitivity whereas the C-terminus has low solubility and is inert to pH changes and vice versa for the MiSp (Minor ampullate spidroin) of Araneus ventricosus.
 Andersson et al., 2017 show how minispidroin can be spun into long fibers
Andersson et al., 2017 show how minispidroin can be spun into long fibers
Herein, part BBa_K4247007 is a composite part formed from the following basic parts: BBa_K4247005 (Minispidroin_NT_N-6His), BBa_K4247001 (Minispidroin_2rep) and BBa_K4247002 (Minispidroin_CT). BBa_K4247007 contains the coding sequence for the full minispidroin protein with 2 repeats of the central repetitive domain and a 6x His-tag in the N-terminus.
Characterization
Optimization of inducer concentration
Aim - To determine the concentration of inducer required for optimal protein expression.
Results - Cell cultures were grown ON at 37°C. Then, the next day, the cultures were diluted to an OD600 of 0.1 and induced with 0.1, 0.3, 0.5 and 1mM IPTG and grew ON. We can clearly see that around 30kDa, there is a darker band in the induced lanes compared to the uninduced lane, showing that the protein is expressed upon induction with IPTG. Further, among the induced lanes, protein expression is maximum when the cultures were induced with 0.3mM IPTG.
Further, a western blot was done on the above SDS-gel to confirm that the proteins we see are indeed the minispidroin proteins. Since the proteins were expressed with a 6x His-tag, we used mouse anti-hexa his primary antibodies and goat anti-mouse HRP-conjugated secondary antibodies for the western blot. Once again, it is clear that 0.3mM IPTG is the optimal inducer concentration.
Conclusion - So, it is clear that 0.3mM IPTG is the optimal concentration for protein expression. This is further backed up by the results of the minispidroin literature since the authors found 0.3mM IPTG to the optimal concentration too.
Optimization of temperature after induction
Aim - To determine the optimal temperature for growing the cells post-induction.
Results - The cultures were grown ON at 37°C. Then, the next day, the cultures were diluted to an OD600 of 0.1 and induced with 0.3mM IPTG and grew ON at 3 different temperatures - 20, 28 and 37°C. Similarly, uninduced controls were grown in identical conditions. The cells either have the 6x His-tag in the N-terminus or the C-terminus.
Conclusion - It can be seen that at all temperatures, the induced cells produce the proteins whereas the uninduced controls don’t and among the induced cultures, protein expression is highest when the cells are incubated at 28°C post-induction. Further, it is clear that more protein is expressed when the 6x His-tag is in N-terminus rather than the C-terminus of the protein.
Optimization of lysis buffer
Aim - To determine the best buffer for cell lysis that provides most of the protein in a soluble state.
Results - In order to lyse our cells by sonication, we used 2 different lysis buffers to decide which lysis buffer gave the most proteins in the soluble fraction. The recipes of the buffers are as follows,
Buffer 1: 50 mM NaH2PO4 + 500 mM sodium chloride + 10 mM imidazole + 0.5% Triton X-100 + 10% glycerol + 2 mM DTT (added fresh, right before use), pH 8.0
Buffer 2: 20mM Tris-Cl, pH 8.0
The cell cultures were centrifuged to obtain the cell pellets which were resuspended in the cell lysis buffer and then sonicated until a clear lysate was obtained. The lysate was centrifuged to obtain the insoluble and soluble fractions in the pellet and supernatant respectively. In the SDS-gel, we can clearly see that most of the protein is in the soluble fraction for both the buffers. Further, it is also clear that we obtain more of the protein in the soluble fraction with buffer 2 which is also the buffer used in the minispidroin literature.
Further, a western blot was done on the above SDS-gel to confirm that the proteins we see are indeed the minispidroin proteins using the above mentioned antibodies. Once again, it is clear that most of the protein is obtained in the soluble fraction with buffer 2.
Conclusion - From the SDS-gel and western blot, we can conclude that buffer 2 is better than buffer 1 since it provides most of the protein in the soluble fraction. This is further backed up by the results of the minispidroin literature since the authors used buffer 2 as well.
Heat purification
Aim - To determine if there is a temperature that would precipitate only the minispidroin proteins without the other E.coli proteins, to facilitate an easy purification method using heat treatment.
Results - The soluble fraction of the lysate was subject to different temperatures (37, 40, 45, 50, 60°C). Then, after heat treatment, the samples were centrifuged and the supernatants and pellets obtained after different temperatures were run on an SDS-gel. It is clear that minispidroin-NT_2rep_CT remains soluble upto 50°C since most of it is found in the supernatant and not the pellet. At 50 and 60°C, most of the protein is found in the pellet showing that it precipitates at temperatures above 50°C. However, a lot of other proteins also precipitate at those temperatures, so it is not possible to obtain the pure protein.
To test if higher temperatures would yield puree proteins, the lysate was subject to 70 and 80°C. It is clear that most of the protein precipitates at 70 and 80°C but like before, it is not very pure since a lot of other proteins are also precipitating.
Further, a western blot was done on the above SDS-gel to confirm that the proteins we see are indeed the minispidroin proteins using the above mentioned antibodies. Once again, it is clear that most of the protein precipitates at 70 and 80°C.
Conclusion - So, although most of the protein precipitates at temperatures above 50°C, heat treatment is not a suitable method for protein purification since there is no temperature that precipitates minispidroin without also precipitating other proteins.
pH purification
Aim - To determine if there is a certain pH at which minispidroin_NT-2rep-CT precipitates without the other E.coli proteins, to facilitate an easy purification method using changes in pH.
Results - The soluble fraction of the lysate was adjusted to different pH (5.5, 6, 6.5, 7, 7.5) and incubated for 1h at room temperature. Then, the samples were centrifuged and the supernatants and pellets obtained at different pH values were run on an SDS-gel. It is clear that minispidroin_NT-2rep-CT does not precipitate at any pH since the lanes with pellets do not have any bands around 30kDa.
Conclusion - Since there is no pH value at which the protein precipitates, the method of adjusting the pH cannot be used for purification.
Optimization of media
Aim - To determine which media, LB (Luria broth) or TB (Terrific broth) is better for protein expression.
Results - The cells were inoculated in either LB or TB media and grown ON at 37°C in identical conditions. The cultures were induced with 0.3mM IPTG and allowed to express the protein ON.
Conclusion - It is clear that more of the protein is expressed when the cells are grown in LB rather than TB.
Protein purification by IMAC
Aim - To purify the protein by IMAC (immobilized metal ion chromatography) using Ni-NTA resin.
Results - Mini columns were loaded with Ni-NTA resin and the soluble fraction of the lysate was added to the columns. Then, the columns were washed twice and eluted to obtain the purified protein. An SDS-gel was run on the different purification fractions and it is clear that most of the protein was lost in the flowthrough and washes and almost nothing was eluted. This shows that the proteins did not bind to the Ni-NTA column at all.
Lysis buffer - 20mM Tris-Cl, pH 8.0
Wash buffer - 20mM Tris-Cl, 5mM Imidazole, pH 8.0
Elution buffer - 20mM Tris-Cl, 200mM Imidazole, pH 8.0
Dialysis buffer - 20mM Tris-Cl, pH 8.0
Since most of the protein was lost in the flowthrough and washes in the previous attempt, the soluble fraction containing the proteins was allowed to incubate with the Ni-NTA resin under shaking conditions for 30-60 mins. This would provide sufficient time for the resin to bind the protein. Then, the columns were washed twice and eluted to obtain the purified protein. An SDS-gel was run on the different purification fractions and it is clear that most of the protein is obtained in the eluate and none of it is lost in the flowthrough or washes. Further, the purity of the obtained protein is high since there are no other bands.
Conclusion - So, with a sufficient incubation time (30-60mins) that allows the Ni-NTA resin to bind all the proteins, it is possible to elute the proteins with very high purity.
A quicker approach to protein purification by IMAC
Aim - To purify the protein by IMAC (immobilized metal ion chromatography) using Ni-NTA resin in a quicker way.
Results - Ni-NTA resin was added to the soluble fraction of the lysate and allowed to incubate for 1h under shaking conditions. Then, it was centrifuged to collect the resin with the proteins and the supernatant was collected as flowthrough. Then, the resin was washed twice and eluted 5 times.
Conclusion - It can be seen that most of the protein is collected in the first elution without much being lost in the flowthrough or the washes.
Stripping the Ni-NTA columns with urea
Aim - To wash the Ni-NTA columns with urea and strip them with EDTA after elution to determine if there was some aggregated protein in the resin or if there were any other metal binding proteins stuck to the column.
Results - After washes and elution, the column was washed with 8M urea. Any aggregated protein that is clumping the column would be solubilized in the urea and hence removed from the column. Then, the column was stripped using EDTA since EDTA can chelate metal ions and hence remove all the Ni and anything bound to it like metal binding proteins. We can see that while some of the protein is lost in the first wash, most of it is in the first elution. Further, a small amount of protein is also seen in the second elution and the urea wash and an even smaller amount after stripping the columns.
Conclusion - Although a small amount of protein is lost in the wash or stuck to the column, most of the protein is obtained in the first elution.
Large scale production
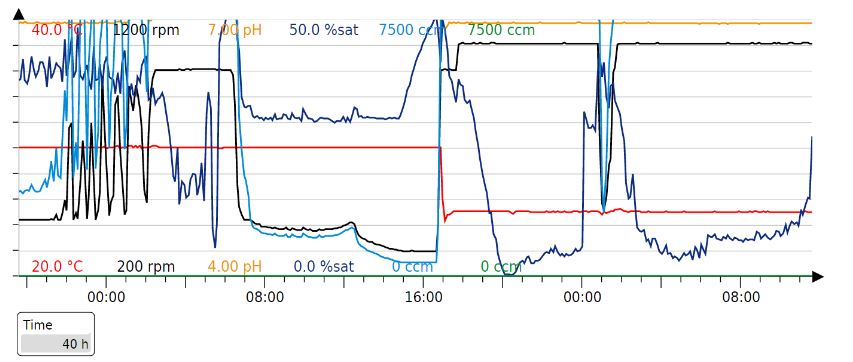
Aim - To carry out large scale protein production by fed-batch fermentation.
Results - In order to obtain large amounts of the protein for spinning the fibre, the proteins were produced in large scale by cultivating the cells in a 1L fermenter. The cells were grown in 1L of batch media at 37°C to an OD600 of 77 which takes about 20h. Then, when the cells have consumed all the media, there is a spike in the oxygen saturation levels due to depletion of glucose. At this point, the cells are induced with 250uM IPTG and fed-batch (500ml) mode is started. Then, after 20h of induction, the fermentation was stopped.
Fermentation data: Settings fermentor from https://doi.org/10.1016/j.mattod.2021.07.020
pO2=30%
T= 37 C growth -> 20 C after induction at around 21h
pH= 7 rpm= 420-1100
total duration= 40h
For 1L of batch media:
Glucose - 14ml of 700g/L stock to achieve 10g/L
MgSO4 - 2.04ml of 245g/L stock to achieve 0.5g/L
Kanamycin - 2ml of 50mg/ml stock to achieve 100mg/L
Metal - 40ml (1:25)
For 1L of fed-batch media:
MgSO4 - 16ml of 245g/L stock to achieve 40g/L
Kanamycin - 2ml of 50mg/ml stock to achieve 100mg/L
Metal - 40ml (1:25)
Metal solutions:
An SDS-gel was done on cells before and after induction.
Then, the cells were lysed and the proteins were purified by IMAC using Ni-NTA resin. Although the purity is not very high as seen by a few other bands in the eluate lane, most of the protein is found in the eluate.
Conclusion - We can see that post-induction, the protein is produced by the cells in the fermenter. Further, purification by Ni-NTA chromatography was successful since a large band can be seen for the protein in the eluate showing that protein production has been increased.
Spinning
Aim - To spin the obtained purified proteins into a fibre.
Results - The purified proteins were dialyzed in a 10kDa cutoff membrane overnight against 20mM Tris-Cl, pH 8.0 to remove all the imidazole. Then, the dialyzed proteins were freeze dried to powder.
The powdered proteins were diluted to 30% w/v with 20mM Tris-Cl, pH 8.0. This concentrated spinning dope was used for spinning the fibre into a coagulation bath - 500 mM sodium acetate buffer, 200 mM NaCl, pH 5. It was extremely difficult to solubilise the powdered proteins and when we tried to spin the dope, the proteins just formed clumps in the coagulation bath as soon as they left the spinning setup.
Since it was extremely difficult to solubilize the powder at 30% w/v, we tried to solubilize the proteins at 15% w/v. This time, it was much easier to solubilize the protein in the buffer. However, when we tried to spin the fibre, the proteins formed clumps in the coagulation bath again and there was no fibre.
Since solubilizing the powdered proteins was difficult, we decided to concentrate the dialyzed proteins directly without freeze drying. We used Amicon 15 centrifugal filter units to concentrate the proteins into a highly concentrated, viscous spinning dope (~30% w/v). The spinning dope was spun into the same coagulation bath using the same spinning setup. As soon as the proteins hit the coagulation bath, they formed long fibres.
Conclusion - Hence, not only was it difficult to solubilize the freeze dried proteins but it was also not possible to spin the fibres from them whereas the proteins that were concentrated without freeze drying could be spun into long fibres.
Protein yields
| None |