Part:BBa_K4409009
igRNA (AAG+GAPDH as guide+ hsa_circ_0001982 as trigger)
This is an RNA-interacting guide RNA (igRNA). It is used in a CRISPR-Cas12 system. It mainly comprises the following three parts. The first part (BBa_K2965036) is the Cas12 handle which combines with Cas12 protein. The second part (BBA_K4409000) is a guide sequence targeting a DNA sequence of GAPDH. The last part (BBa_K4409004) is a control sequence made up of VR1, loop and VR2 regions in sequence. VR1 and VR2 regions can specifically bind to the RNA trigger which is a unique sequence of hsa_circ_0001982. The loop binds to a portion of the guide sequence. When hsa_circ_0001982 is present in the CRISPR-Cas12 system, the control sequence binds with it and the igRNA is activated. The loop then unblocks the guide. The guide binds to a GAPDH dsDNA with the PAM sequence, and activates the Cas12 protein, resulting in the cleavage of dsDNA and ssDNA in the system.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Results
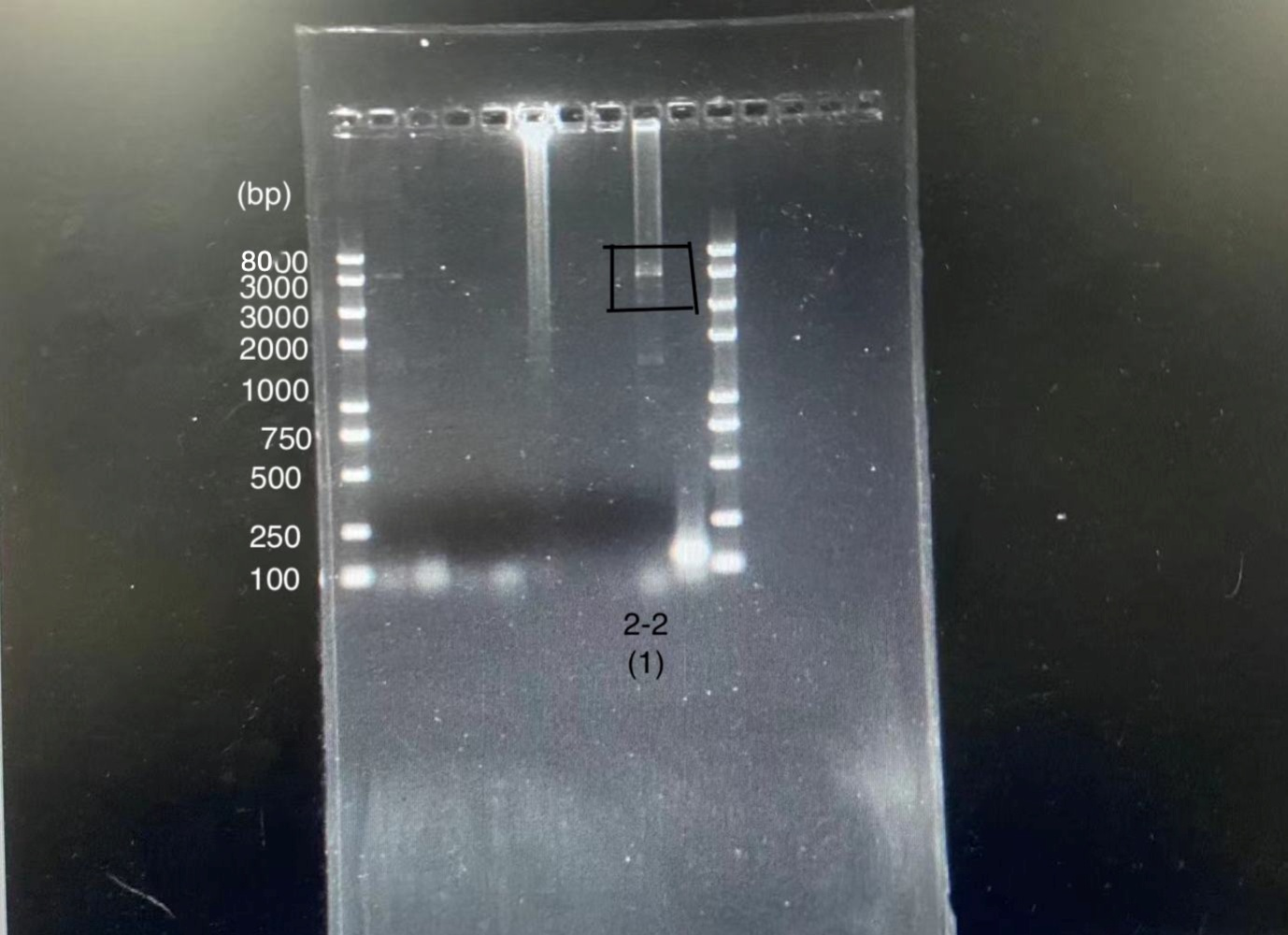
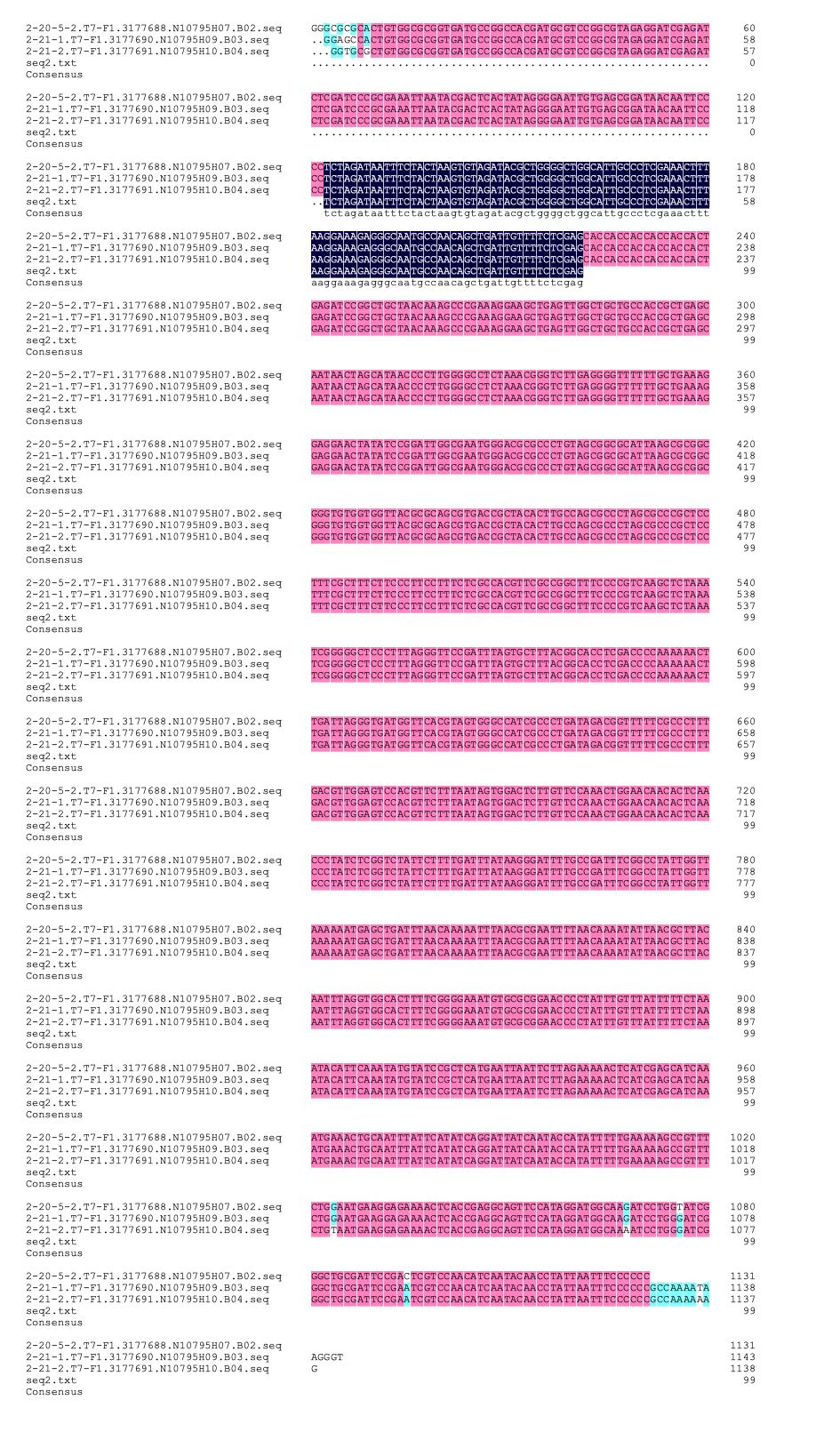
We constructed a plasmid based on pET-28a plasmid by inserting the sequence of this part between the restriction enzyme sites using t-PCR method. PCR results showed the plasmid was constructed successfully since the site of the band is between 5000bp and 8000bp, shown in Graph 2, and the length of plasmid we constructed was also in this region, exactly 5455bp. Moreover, we confirmed the sequence by gene sequencing, shown in Graph 3. All of the base pairs in the main region turns out to be accurate.
A CRISPR-Cas12 system using this igRNA produced fluorescence significantly bigger than negative control groups. The fluorescence produced is shown in graph 4 and table 1-2.
| None |