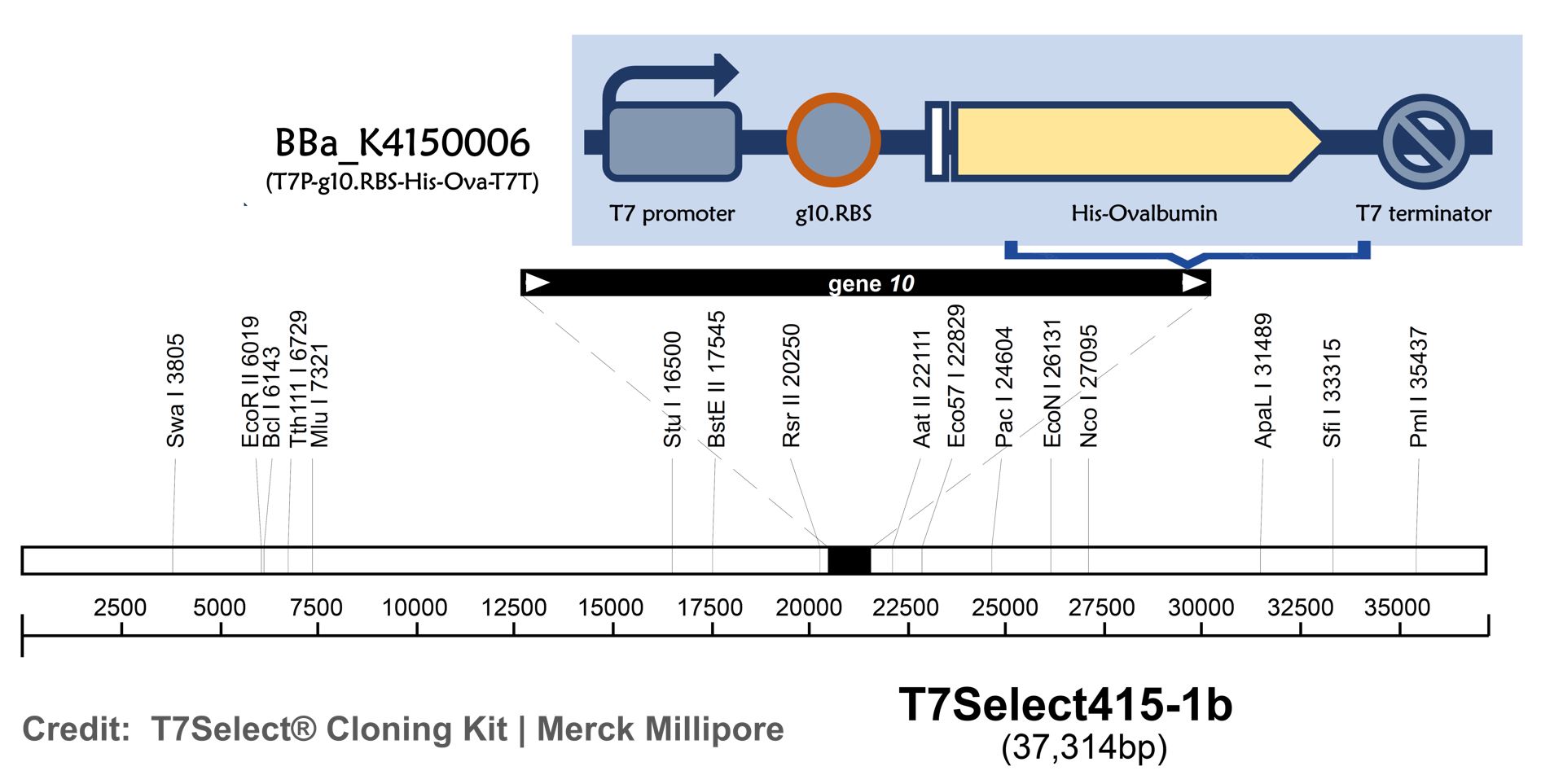
Part:BBa_K4150006
T7P-g10.RBS-His-Ova-T7T
ThisThe chicken ovalbumin (OVA) is a glycoprotein, which is a major component of chicken egg whites, and harbors immunogenic properties in vaccination experiments[1]. The recombinant OVA protein is readily purified in E. coli BL21 system driven by T7 promoter and triggered by IPTG induction. Therefore, it’s considered as one of the model antigens used to study immune responses in animal models.
ThisTo engineer bacteriophage vaccine vector, we chose a T7 phage-based system because it’s one of the well-studied model bacteriophages with rich lab materials and available assay methods such as T7 promoter, IPTG induction in E. coli BL21 strain, in vitro transcription and translation (TXTL) studies, phage genome sequences and engineering tools[2] as well as safe application in clinical trials[3].
Engineering of Bacteriophage
Plasmid Construction
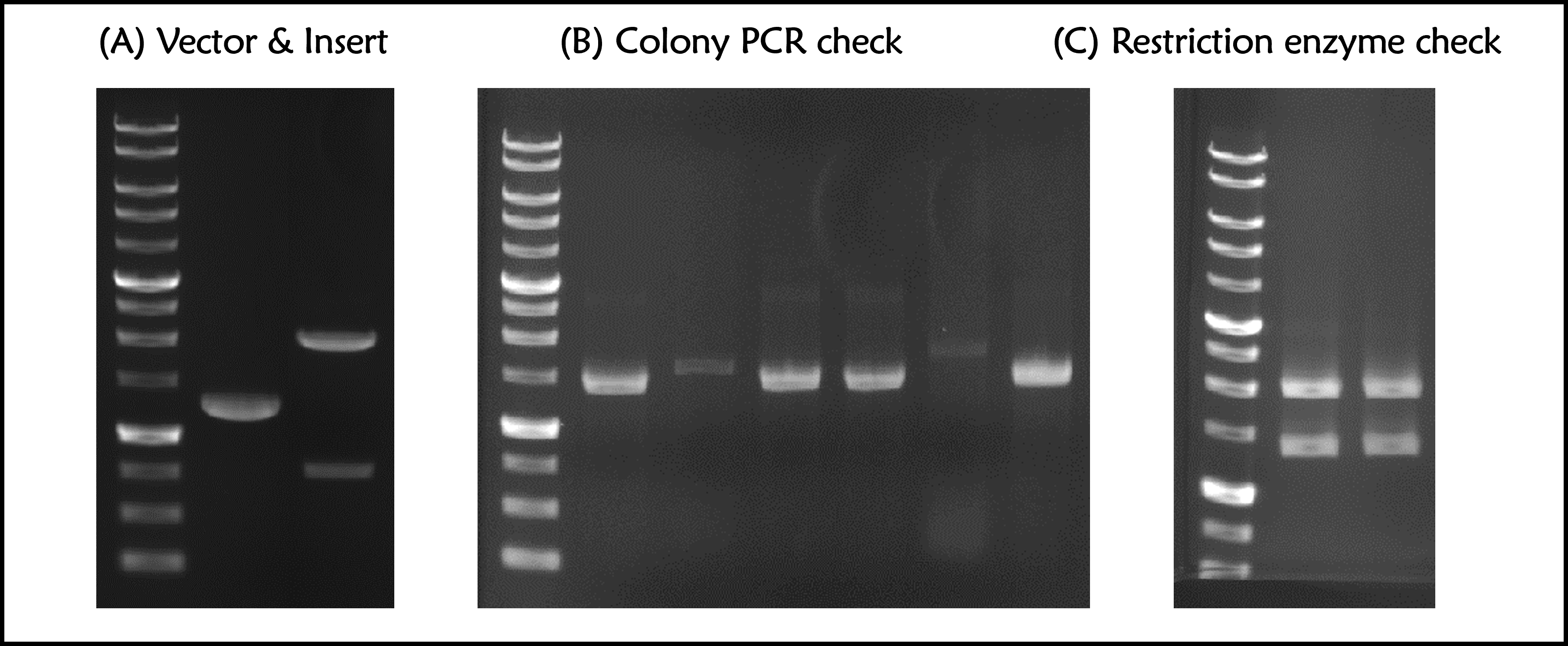
ThisPrior to engineer T7 bacteriophage, we constructed a composite biobrick part composed of T7 promoter from an existing part, a novel enhanced RBS with a g10 leader sequence, Gallus gallus (Chicken) ovalbumin antigen gene with a 6xHis tag, and a T7 terminator. The vector was prepared from Part:BBa_K4150005 as a backbone with a T7 promoter and a T7 terminator by digested with SpeI and HindIII. The insert of g10.RBS-His-Ova was synthesized by Integrated DNA Technologies, Inc., followed by digested with XbaI and HindIII. E. coli DH5alpha competent cells were transformed with the resulting ligation product and checked by colony PCR and restriction enzymes, as well as confirmed by DNA sequencing.
Figure 1 | T7P-g10.RBS-His-Ova-T7T (Part:BBa_K4150006) construction. DNAs were run electrophoresis on 1% agarose gel with 1kb marker. (A) Vector (2148 bp with a 835-bp DNA fragment after cut) and Insert (g10.RBS-His-Ova, 1285 bp) were digested by indicated restriction enzymes before ligation. (B) 6 colonies were subject to colony PCR with VF2 forward primer and Ova reverse primers (PCR product size: 1449 bp). (C) The 2 of plasmid DNAs with a correct PCR product size were extracted and digested by EcoRI and PstI (Product size: 2029 and 1398 bps).
Phage Engineering
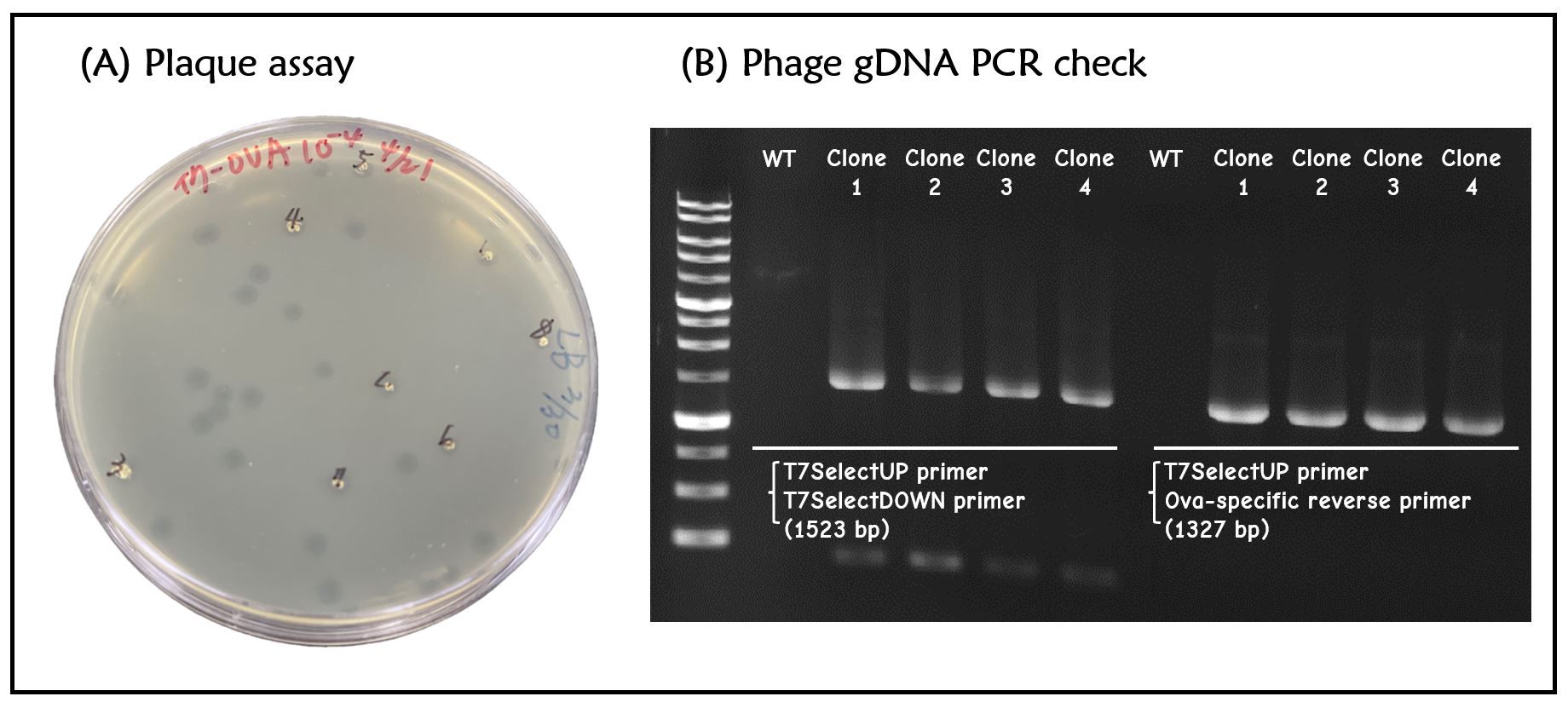
ThisT7 bacteriophage genome was edited with the BioBrick of T7P-g10.RBS-His-Ova-T7T in the end of T7 gene 10 using the T7Select® 415-1 Cloning Kit (Merck Millipore) according to the manufacturer’s instruction. To select the correct T7 phage genome inserted with the BioBrick, the plaque assay on LB agar plate covered by the lawn of E. coli DH5alpha was performed. Several phages from the plaques were subjected to phage genomic DNA extraction and PCR with corresponding primers (Fig. 2).
Figure 2 | T7 phage engineering with T7P-g10.RBS-His-Ova-T7T. (A) The plaque assay was conducted on the LB agar plate, where E. coli DH5alpha cells were infected with the engineered phages. (B) The gDNAs of phages from 4 plaques along with wild-type T7 phages were extracted and subjected to PCR with the indicated primer pairs. PCR product sizes were shown in the figure. DNAs were run electrophoresis on 1% agarose gel with 1kb marker.
Characterization of the Elements
T7 promoter, RBS & T7 terminator
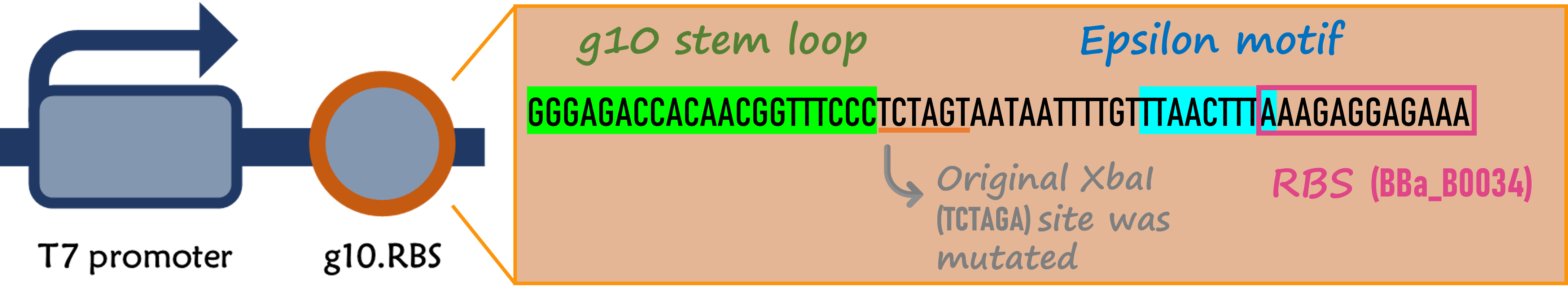
Enhanced g10.RBS with a T7 gene 10 leader sequence
ThisIn order to increase the expression of protein of interest, we added a canonical strong RBS (Part:BBa_B0034) with a leader sequence with a stem loop from the T7 bacteriophage gene 10 (g10.RBS, Part:BBa_K4150000).
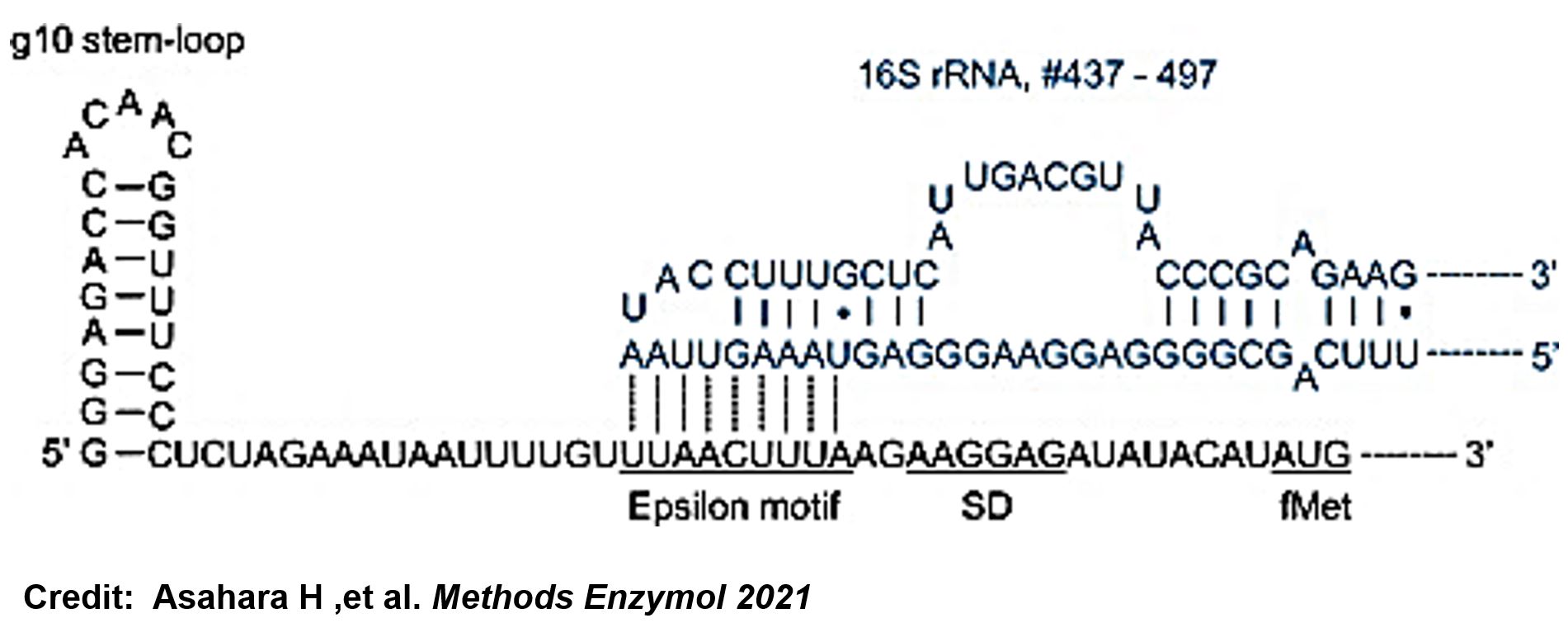
ThisThe g10 leader sequence encodes very highly expressed phage proteins, in which 9-base of Epsilon motif exhibits perfect complementary to the 16S RNA of E. coli[4]. This structure connected with the consensus Shine-Dalgarno sequence (SD) can enhance up to 340-fold heterologous gene expression in E. coli[5]. The g10 leader sequence is also present in several commercially available pET series vectors.
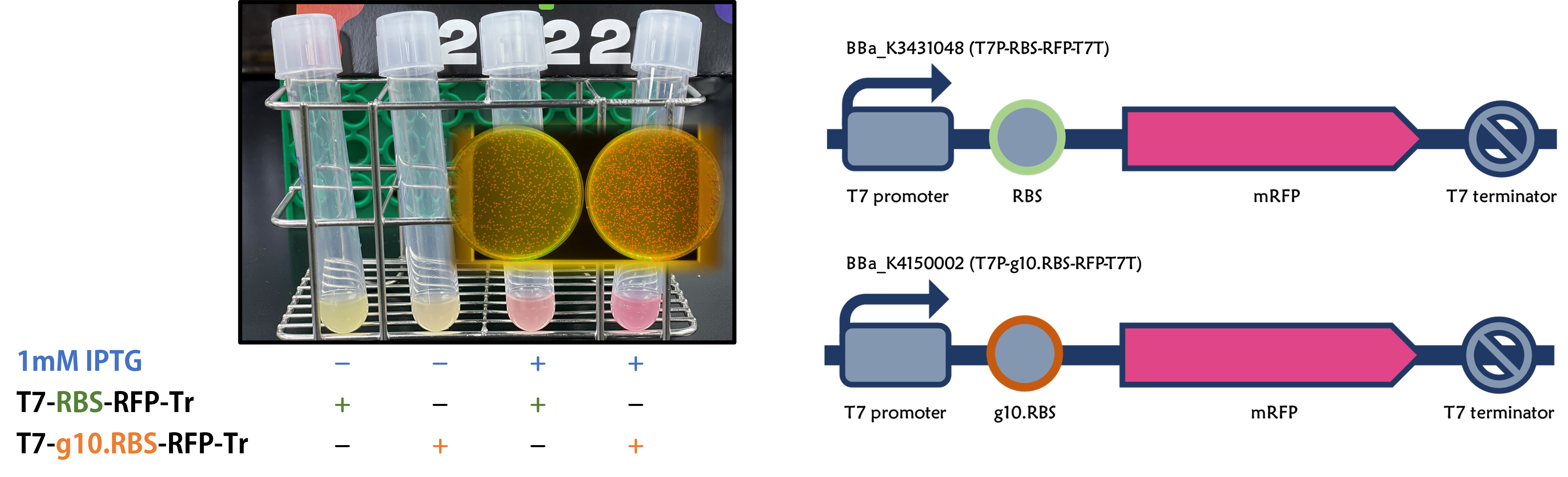
ThisWe replaced the RBS with g10.RBS in BioBrick Part:BBa_K3431048 (T7-RBS-RFP-Tr) to create an improved BioBrick Part:BBa_K4150002 (T7-g10.RBS-RFP-Tr). These two plasmids were transformed into E. coli BL21. In the presence of IPTG, colonies on the LB agar plate and cultures in the LB broth both showed darker red colors for RFP gene expression driven by T7 promoter with g10 leader sequence compared to the original RBS without g10 leader sequence (Fig.1). However, the E. coli BL21 with T7-g10.RBS-RFP-Tr had a higher background level in the absence of IPTG.
Figure 1 | The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown on LB agar plate supplemented with 20 μg/mL of chloramphenicol and 1mM of IPTG. One of the colonies was cultured in LB broth with 34 μg/mL of chloramphenicol in the absence or in the presence of 1mM of IPTG. Inset panel: The plates were observed under a blue LED light box.
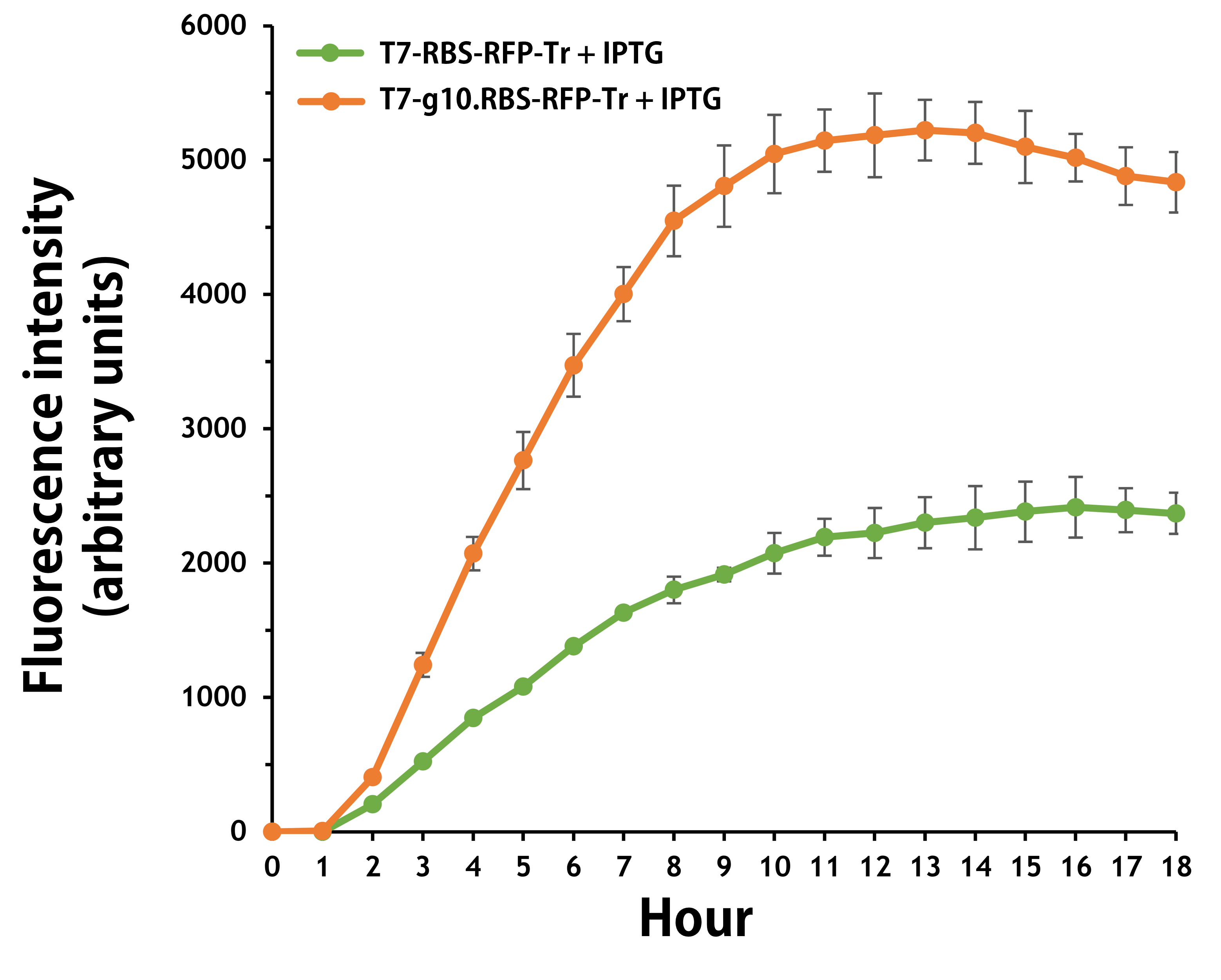
ThisFurthermore, the transformed E. coli with T7-g10.RBS-RFP-Tr compared to T7-RBS-RFP-Tr expressed RFP at faster and increased (up to 2.5-fold) fluorescence intensity levels in a time-dependent manner (Fig. 2). The above data demonstrated the g10 leader sequence in front of RBS can significantly enhance gene expression.
Figure 2 | The plasmids as indicated were transformed into E. coli BL21. The E. coli were grown in LB broth supplemented with 34 μg/mL of chloramphenicol and 1mM of IPTG for 18 hours. The RFP expression levels were read at ex/em = 584/607 nm in a kinetic mode by Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments (Agilent Technologies, Inc.). The values of fluorescence intensity were presented by the data with IPTG induction minus the data without induction as background levels.
Engineering of Bacteriophage
Reporter - T7 phage::GFP
Vaccine - T7 phage::OVA
Reference
- ↑ Geary TW, Reeves JJ. Production of a genetically engineered inhibin vaccine. Vaccine. 1996 Sep;14(13):1273-9. doi: 10.1016/s0264-410x(96)00014-x. PMID: 8961517.
- ↑ Ludwig T, Hoffmann R, Krizsan A. Construction and Characterization of T7 Bacteriophages Harboring Apidaecin-Derived Sequences. Curr Issues Mol Biol. 2022 Jun 1;44(6):2554-2568. doi: 10.3390/cimb44060174. PMID: 35735615; PMCID: PMC9221748.
- ↑ McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, Brüssow H. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology. 2013 Sep 1;443(2):187-96. doi: 10.1016/j.virol.2013.05.022. Epub 2013 Jun 10. PMID: 23755967.
- ↑ Olins PO, Devine CS, Rangwala SH, Kavka KS. The T7 phage gene 10 leader RNA, a ribosome-binding site that dramatically enhances the expression of foreign genes in Escherichia coli. Gene. 1988 Dec 15;73(1):227-35. doi: 10.1016/0378-1119(88)90329-0. PMID: 3072257.
- ↑ Asahara H, Magnelli P, Shi X, Tuckey C, Zhou Y, Samuelson JC. Guidelines for nucleic acid template design for optimal cell-free protein synthesis using an Escherichia coli reconstituted system or a lysate-based system. Methods Enzymol. 2021;659:351-369. doi: 10.1016/bs.mie.2021.07.005. Epub 2021 Sep 2. PMID: 34752294.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1315
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 34
Illegal SapI.rc site found at 468
| None |