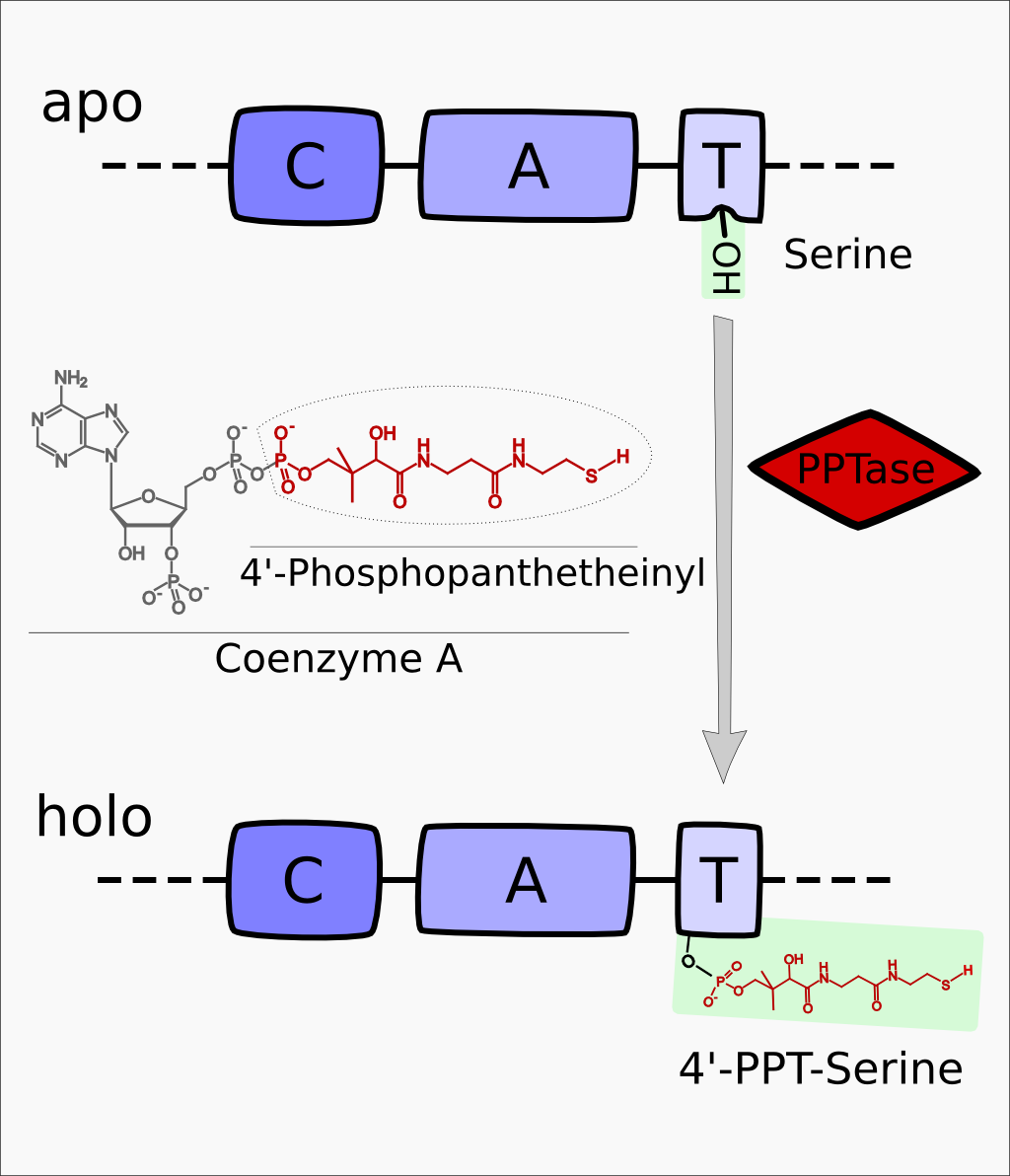
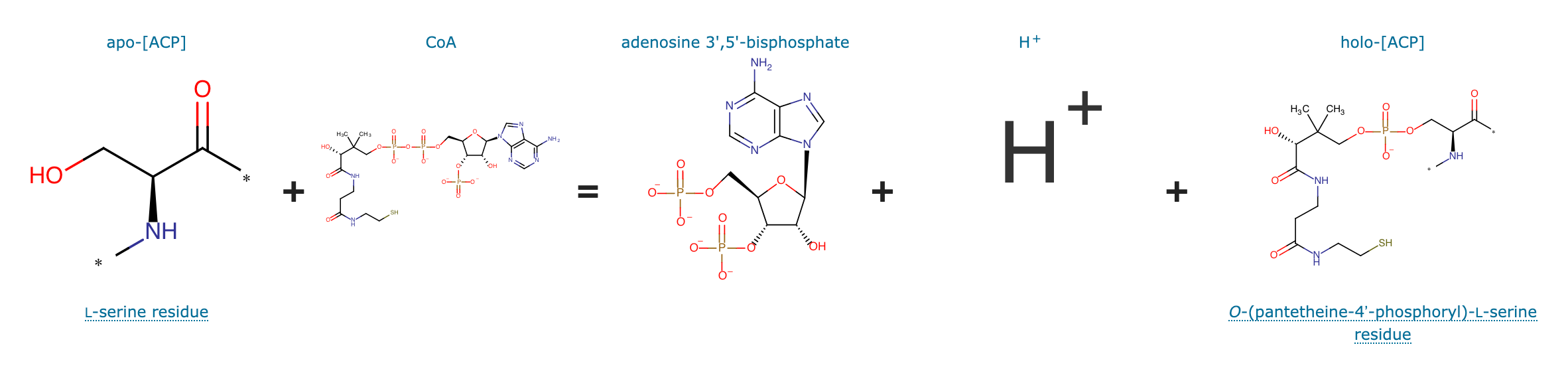
Part:BBa_K302010
Svp 4'-Phosphopanthetheinyl-transferase
svp encodes for a 4'-Phosphopanthetheinyl-transferase (PPTase) which transfers a 4'-PPT residue from CoA to a conserved serine residue in the T-Domain of NRPS modules, thus activating these enzymes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 173
Illegal NgoMIV site found at 413
Illegal NgoMIV site found at 663
Illegal AgeI site found at 573 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 366
References
- Takahashi, H., Kumagai, T., Kitani, K., Mori, M., Matoba, Y., & Sugiyama, M. (2007). Cloning and Characterization of a Streptomyces Single Module Type Non-ribosomal Peptide Synthetase Catalyzing a Blue Pigment Synthesis *, 282(12), 9073–9081. doi:10.1074/jbc.M611319200
Information contributed by City of London UK (2021)
Part information is collated here to help future users of the BioBrick registry.
Metadata:
- Group: City of London UK 2021
- Author: Janusan Jeyananthan
- Summary: Added information collated from existing scientific studies
Kinetics:
kcat is 86 min(-1) with S.glaucescens apo-ACP TcmM as the substrate and 11 min(-1) with S.mobaraensis apo-PCP BlmI as the substrate.
For Streptomyces mobaraensis apo-PCP BlmI1: KM=3.9 µM For Streptomyces glaucescens apo-ACP TcmM: KM=3.1 µM
References
Information contributed by NFLS_Nanjing (2022)
Part information is collated here to help future users of the BioBrick registry.
Metadata:
- Group: NFLS_Nanjing (2022)
- Author: Yuming Xue
- Summary: Added information collated from existing scientific studies
Biology and usage:
1. The gene bpsA is expressed in C. glutamicum and carries strong fluxes toward L-glutamate, a precursor of indigoidine. [1]
2. The gene bpsA catalyzes indigoidine formation from two molecules of glutamine in an ATP-dependent manner.[2]
Experimental approach:
1. Integrated sfp into the yeast chromosomal δ-sequences.
2. Codon-optimize Te bpsA gene for expression in S. cerevisiae and genomically integrated into locus ARS1014a under control of the TDH3 promoter and ADH1 terminator using a previously reported, cloning free Cas9 toolkit.
3. Use the conventional lithium acetate method to perform transformations.
4. Use 200 ng pCut_1014a and 500 ng of linear Donor DNA with 500 bp homology to the integration locus ARS1014a.[3]
References
[1] High-Level Production of the Natural Blue Pigment Indigoidine from Metabolically Engineered Corynebacterium glutamicum for Sustainable Fabric Dyes [2] Genome-scale metabolic rewiring improves titersrates and yields of the non-native product indigoidine at scale
[3] Production efciency of the bacterial non-ribosomal peptide indigoidine relies on the respiratory metabolic state in S. cerevisiae- ↑ “Svp - 4’-Phosphopantetheinyl Transferase Svp - Streptomyces Mobaraensis - Svp Gene & Protein” n.d. Uniprot. Accessed 12 August, 2021 https://www.uniprot.org/uniprot/Q9F0Q6
| None |